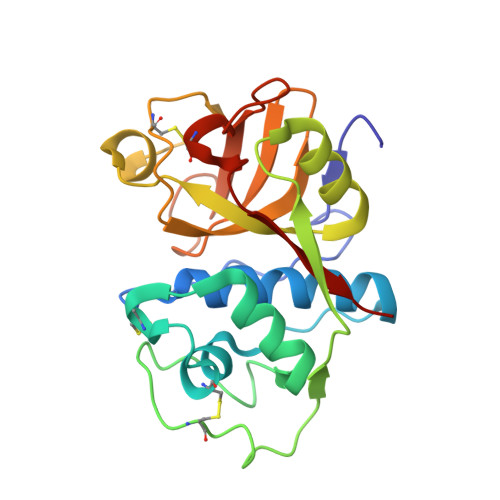
Structure of the mexicain-E-64 complex and comparison with other cysteine proteases of the papain family.
Gavira, J.A., Gonzalez-Ramirez, L.A., Oliver-Salvador, M.C., Soriano-Garcia, M., Garcia-Ruiz, J.M.(2007) Acta Crystallogr D Biol Crystallogr 63: 555-563
- PubMed: 17452780
- DOI: https://doi.org/10.1107/S0907444907005616
- Primary Citation of Related Structures:
2BDZ - PubMed Abstract:
Mexicain is a 23.8 kDa cysteine protease from the tropical plant Jacaratia mexicana. It is isolated as the most abundant product after cation-exchange chromatography of the mix of proteases extracted from the latex of the fruit. The purified enzyme inhibited with E-64 [N-(3-carboxyoxirane-2-carbonyl)-leucyl-amino(4-guanido)butane] was crystallized by sitting-drop vapour diffusion and the structure was solved by molecular replacement at 2.1 A resolution and refined to an R factor of 17.7% (R(free) = 23.8%). The enzyme belongs to the alpha+beta class of proteins and the structure shows the typical papain-like fold composed of two domains, the alpha-helix-rich (L) domain and the beta-barrel-like (R) domain, separated by a groove containing the active site formed by residues Cys25 and His159, one from each domain. The four monomers in the asymmetric unit show one E-64 molecule covalently bound to Cys25 in the active site and differences have been found in the placement of E-64 in each monomer.
- Laboratorio de Estudios Cristalográficos, IACT, CSIC-Universidad de Granada, Granada, Spain.
Organizational Affiliation: