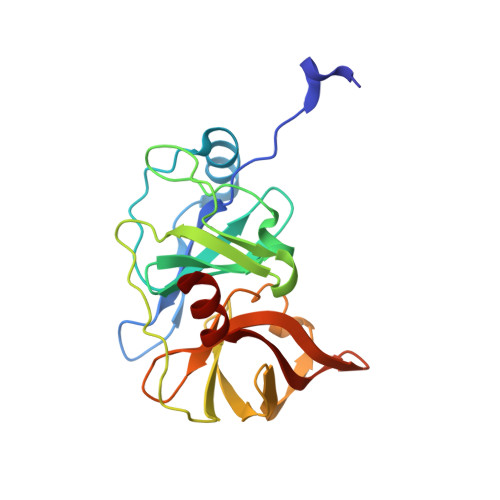
Molecular mechanism of pan-genotypic HCV NS3/4A protease inhibition by glecaprevir and characterization of genotype-specific structural differences
Timm, J., Kosovrasti, K., Henes, M., Leidner, F., Hou, S., Kurt-Yilmaz, N., Schiffer, C.A.To be published.