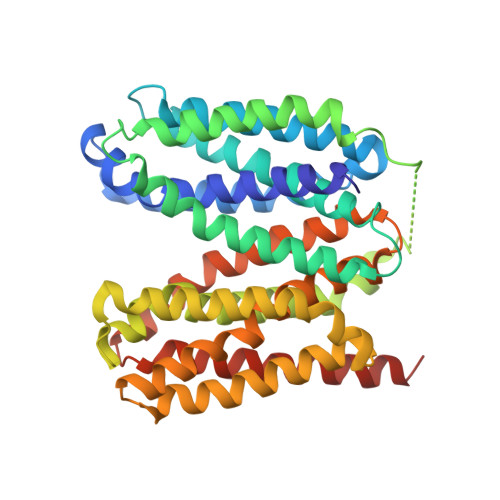
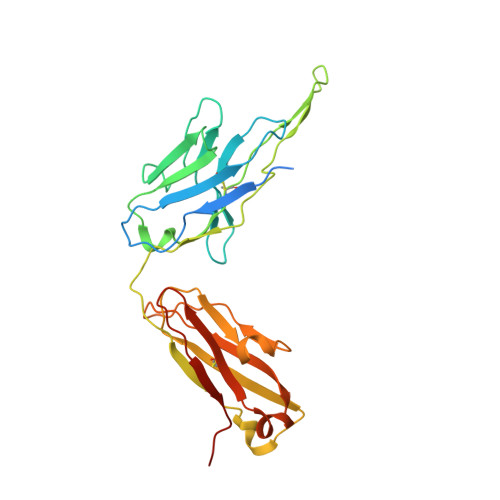
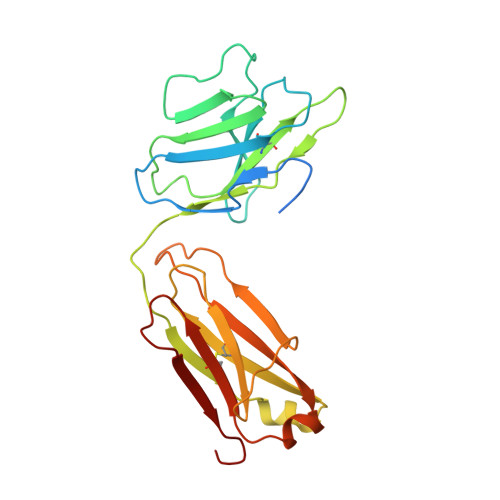
Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus.
Brawley, D.N., Sauer, D.B., Li, J., Zheng, X., Koide, A., Jedhe, G.S., Suwatthee, T., Song, J., Liu, Z., Arora, P.S., Koide, S., Torres, V.J., Wang, D.N., Traaseth, N.J.(2022) Nat Chem Biol 18: 706-712
- PubMed: 35361990
- DOI: https://doi.org/10.1038/s41589-022-00994-9
- Primary Citation of Related Structures:
7LO7, 7LO8 - PubMed Abstract:
Membrane protein efflux pumps confer antibiotic resistance by extruding structurally distinct compounds and lowering their intracellular concentration. Yet, there are no clinically approved drugs to inhibit efflux pumps, which would potentiate the efficacy of existing antibiotics rendered ineffective by drug efflux. Here we identified synthetic antigen-binding fragments (Fabs) that inhibit the quinolone transporter NorA from methicillin-resistant Staphylococcus aureus (MRSA). Structures of two NorA-Fab complexes determined using cryo-electron microscopy reveal a Fab loop deeply inserted in the substrate-binding pocket of NorA. An arginine residue on this loop interacts with two neighboring aspartate and glutamate residues essential for NorA-mediated antibiotic resistance in MRSA. Peptide mimics of the Fab loop inhibit NorA with submicromolar potency and ablate MRSA growth in combination with the antibiotic norfloxacin. These findings establish a class of peptide inhibitors that block antibiotic efflux in MRSA by targeting indispensable residues in NorA without the need for membrane permeability.
- Skirball Institute of Biomolecular Medicine, New York University School of Medicine, New York, NY, USA.
Organizational Affiliation: