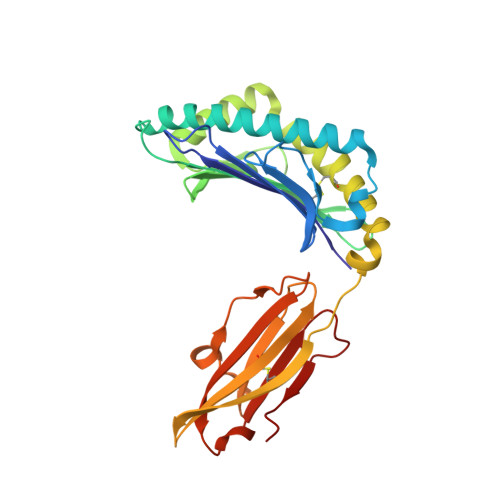
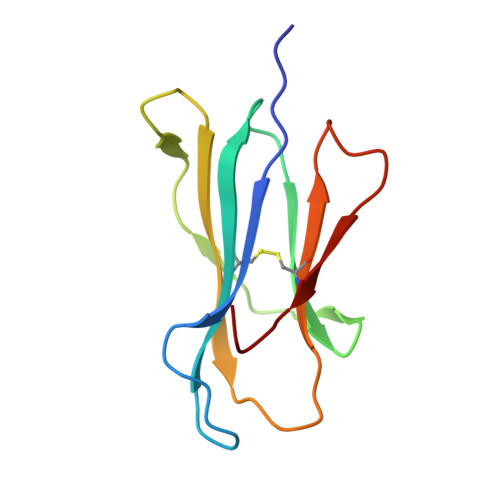
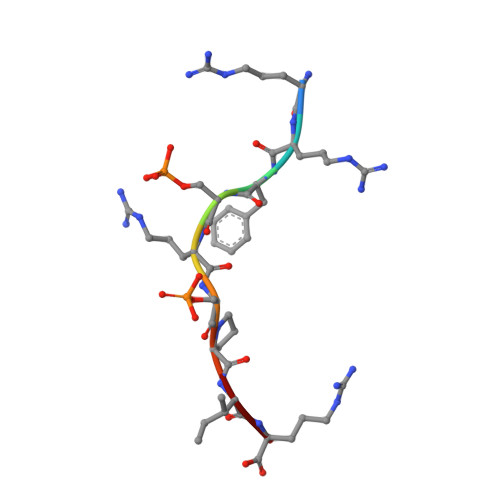
Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Zhao, Y., Sun, M., Zhang, N., Liu, X., Yue, C., Feng, L., Ji, S., Liu, X., Qi, J., Wong, C.C.L., Gao, G.F., Liu, W.J.(2022) iScience 25: 104013-104013
- PubMed: 35310951
- DOI: https://doi.org/10.1016/j.isci.2022.104013
- Primary Citation of Related Structures:
7CIQ, 7CIR, 7CIS, 7DYN - PubMed Abstract:
Phosphopeptides presented by major histocompatibility complex (MHC) class I have been regarded as a pivotal type of cancer neoantigens that are recognized by T cells. The structural basis of single-phosphorylated peptide presentation has been well studied. Diphosphorylation with one interval between two sites is one of the prevalent forms of multisite-phosphorylated peptides. Herein, we determined the molecular basis of presentation of two P4/P6 double pS-containing peptides by HLA-B27 and compared them with unmodified and single-phosphorylated peptide complexes. These data clarified not only the HLA allele-specific presentation of phosphopeptides by MHC class I molecules but also the cooperativity of peptide conformation within P4 and P6 phosphorylation sites. The phosphorylation of P6 site can influence the binding mode of P4 phosphorylated site to HLA-B27. And we found the diphospho-dependent attenuated effect of peptide binding affinity. This study provides insights into the MHC presentation features of diphosphopeptides, which is different from monophosphopeptides.
- NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention (China CDC), Beijing 100052, China.
Organizational Affiliation: