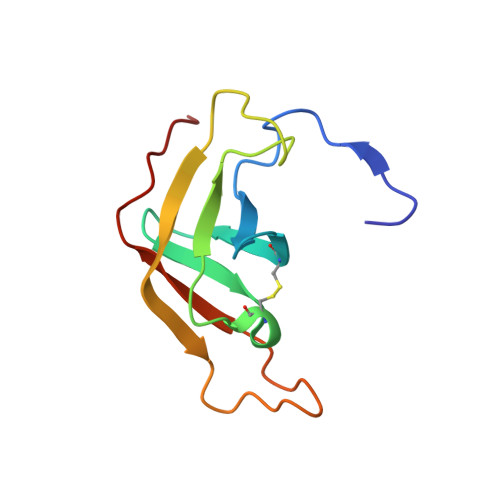
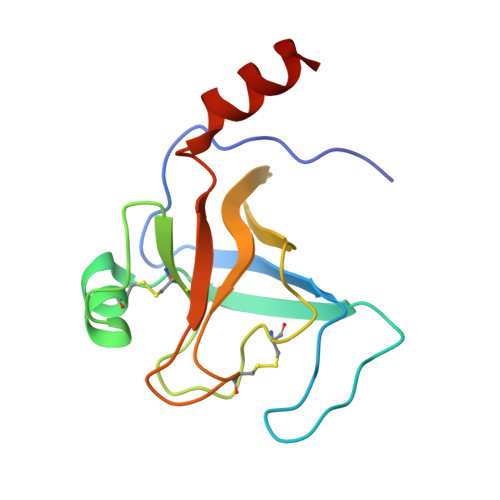
Structural Basis of the Pancreatitis-Associated Autoproteolytic Failsafe Mechanism in Human Anionic Trypsin.
Nagel, F., Susemihl, A., Geist, N., Mohlis, K., Palm, G.J., Lammers, M., Delcea, M.(2022) J Inflamm Res 15: 3633-3642
- PubMed: 35775010
- DOI: https://doi.org/10.2147/JIR.S367699
- Primary Citation of Related Structures:
7Z9F - PubMed Abstract:
The pathophysiological mechanisms underlying chronic pancreatitis (CP) are still poorly understood. Human cationic (TRY1) and anionic (TRY2) trypsins are the two major trypsin isoforms and their activities are tightly regulated within pancreatic acinar cells. Typically, they exist in a molar ratio of 2:1 (cationic:anionic). This ratio is reversed during chronic alcohol abuse, pancreatic cancer, or pancreatitis due to selectively upregulated expression of TRY2, causing anionic trypsin to become the predominant isoform. The involvement of TRY2 in pancreatitis is considered limited due to the absence of disease-causing mutations and its increased prevalence for autoproteolysis. However, exacerbated pancreatitis in TRY2 overexpressing mice was recently demonstrated. Here, we aim to elucidate the molecular structure of human anionic trypsin and obtain insights into the autoproteolytic regulation of tryptic activity. Trypsin isoforms were recombinantly expressed in E. coli , purified and refolded. Enzymatic activities of all trypsin isoforms were determined and crystals of TRY2 were grown using the vapor-diffusion method. The structure was solved by molecular replacement and refined to a resolution of 1.7 Å. Equilibration molecular dynamics simulations were used to generate the corresponding TRY1-TRY1 model. All trypsin isoforms display similar kinetic properties. The crystal structure of TRY2 reveals that the enzyme crystallized in the autoproteolytic state with Arg122 placed in the S1 binding pocket and the corresponding loop cleaved. The TRY2-TRY2 dimer confirms a previously hypothesized autoinhibitory state with an unexpectedly large binding interface. We provide a structure of TRY2, which is the predominant trypsin isoform in chronic pancreatitis and pancreatic cancer. A proposed autoinhibition mode was confirmed and the structural basis of the autoproteolytic failsafe mechanism elucidated.
- Biophysical Chemistry, Institute of Biochemistry, University of Greifswald, Greifswald, Germany.
Organizational Affiliation: