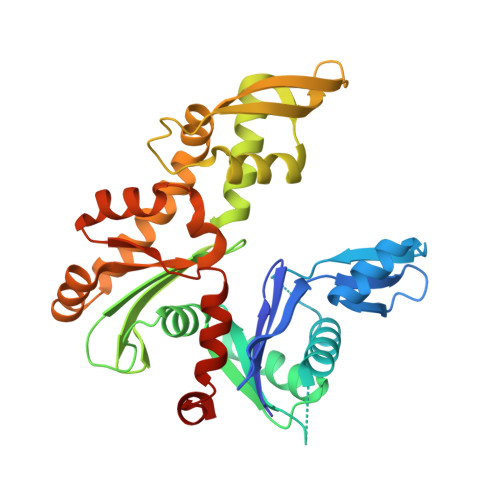
Polymerization cycle of an actin homolog MreB from a Gram-positive bacterium
Mao, W., Conilleau, C., Li de la Sierra-Gallay, I., Ah-Seng, Y., van Tilbeurgh, H., Renner, L., Nessler, S., Bertin, A., Chastanet, A., Carballido-Lopes, R.To be published.