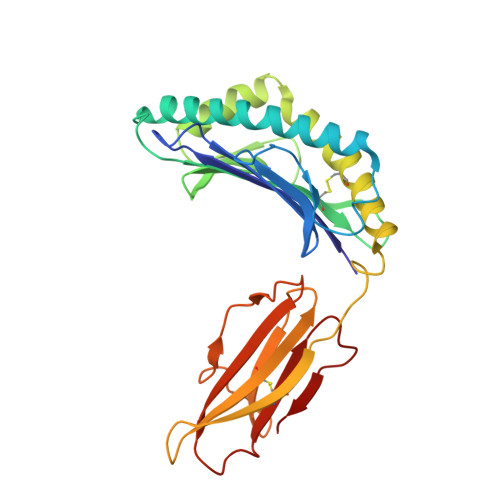
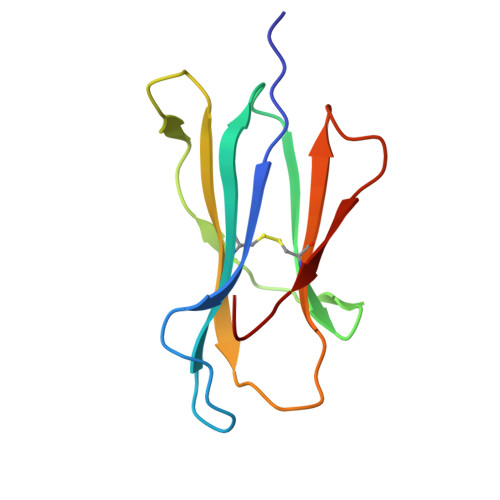
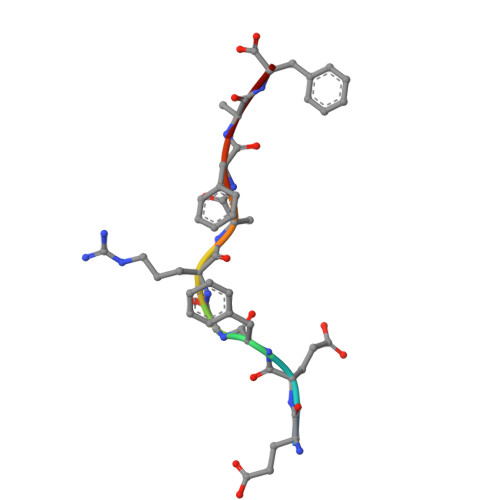
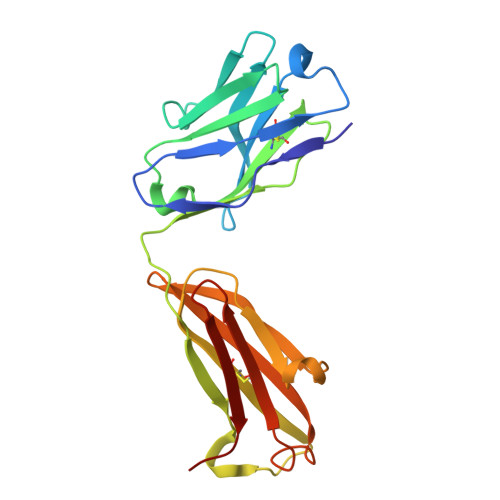
Structural mechanism of anti-MHC-I antibody blocking of inhibitory NK cell receptors in tumor immunity.
Jiang, J., Panda, A.K., Natarajan, K., Lei, H., Sharma, S., Boyd, L.F., Towler, R.R., Chempati, S., Ahmad, J., Morton, A.J., Lang, Z.C., Sun, Y., Sgourakis, N., Meier-Schellersheim, M., Huang, R.K., Shevach, E.M., Margulies, D.H.(2026) Commun Biol
- PubMed: 41629525
- DOI: https://doi.org/10.1038/s42003-026-09641-8
- Primary Citation of Related Structures:
8TQ6, 9D72, 9D73, 9D74, 9OA9 - PubMed Abstract:
Anti-major histocompatibility complex class I (MHC-I) mAbs can stimulate immune responses to tumors and infections by blocking suppressive signals delivered via various immune inhibitory receptors. To understand such functions, we determined the structure of a highly cross-reactive anti-human MHC-I mAb, B1.23.2, in complex with the MHC-I molecule HLA-B*44:05 by both cryo-electron microscopy (cryo-EM) and X-ray crystallography. Structural models determined by the two methods were essentially identical revealing that B1.23.2 binds a conserved region on the α2 1 helix that overlaps the killer immunoglobulin-like receptor (KIR) binding site. Structural comparison to KIR/HLA complexes reveals a mechanism by which B1.23.2 blocks inhibitory receptor interactions, leading to natural killer (NK) cell activation. B1.23.2 treatment of the human KLM-1 pancreatic cancer model in humanized (NSG-IL15) mice provides evidence of suppression of tumor growth. Such anti-MHC-I mAb that block inhibitory KIR/HLA interactions may prove useful for tumor immunotherapy.
- Molecular Biology Section, Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA. jiangji@niaid.nih.gov.
Organizational Affiliation: