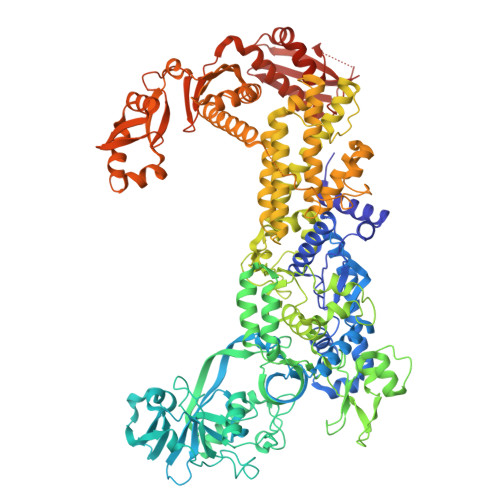
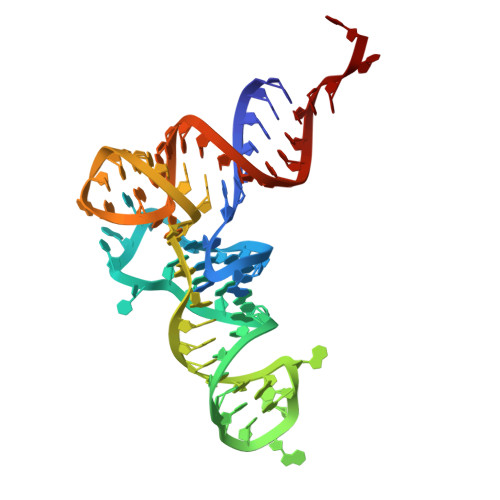
The mechanism of discriminative aminoacylation by isoleucyl-tRNA synthetase based on wobble nucleotide recognition.
Chen, B., Yi, F., Luo, Z., Lu, F., Liu, H., Luo, S., Gu, Q., Zhou, H.(2024) Nat Commun 15: 10817-10817
- PubMed: 39738040
- DOI: https://doi.org/10.1038/s41467-024-55183-0
- Primary Citation of Related Structures:
8WND, 8Z1P - PubMed Abstract:
The faithful charging of amino acids to cognate tRNAs by aminoacyl-tRNA synthetases (AARSs) determines the fidelity of protein translation. Isoleucyl-tRNA synthetase (IleRS) distinguishes tRNA Ile from tRNA Met solely based on the nucleotide at wobble position (N34), and a single substitution at N34 could exchange the aminoacylation specificity between two tRNAs. Here, we report the structural and biochemical mechanism of N34 recognition-based tRNA discrimination by Saccharomyces cerevisiae IleRS (ScIleRS). ScIleRS utilizes a eukaryotic/archaeal-specific arginine as the H-bond donor to recognize the common carbonyl group (H-bond acceptor) of various N34s of tRNA Ile , which induces mutual structural adaptations between ScIleRS and tRNA Ile to achieve a preferable editing state. C34 of unmodified tRNA Ile (CAU) (behaves like tRNA Met ) lacks a relevant H-bond acceptor, which disrupts key H-bonding interactions and structural adaptations and suspends the ScIleRS·tRNA Ile (CAU) complex in an initial non-reactive state. This wobble nucleotide recognition-based structural adaptation provides mechanistic insights into selective tRNA aminoacylation by AARSs.
- State Key Laboratory of Anti-Infective Drug Discovery and Development, School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou, 510006, China.
Organizational Affiliation: