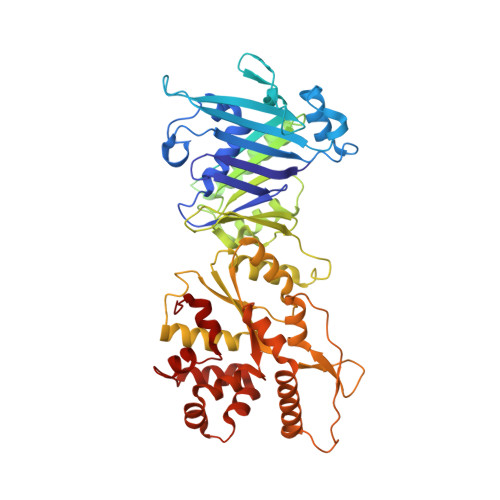
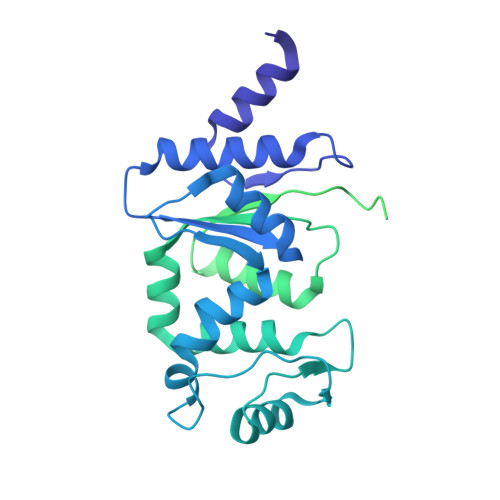
Structures and activation mechanism of the Gabija anti-phage system.
Li, J., Cheng, R., Wang, Z., Yuan, W., Xiao, J., Zhao, X., Du, X., Xia, S., Wang, L., Zhu, B., Wang, L.(2024) Nature 629: 467-473
- PubMed: 38471529
- DOI: https://doi.org/10.1038/s41586-024-07270-x
- Primary Citation of Related Structures:
8JQ9, 8JQB, 8JQC, 8WY4, 8WY5, 8X51, 8X5I, 8X5N - PubMed Abstract:
Prokaryotes have evolved intricate innate immune systems against phage infection 1-7 . Gabija is a highly widespread prokaryotic defence system that consists of two components, GajA and GajB 8 . GajA functions as a DNA endonuclease that is inactive in the presence of ATP 9 . Here, to explore how the Gabija system is activated for anti-phage defence, we report its cryo-electron microscopy structures in five states, including apo GajA, GajA in complex with DNA, GajA bound by ATP, apo GajA-GajB, and GajA-GajB in complex with ATP and Mg 2+ . GajA is a rhombus-shaped tetramer with its ATPase domain clustered at the centre and the topoisomerase-primase (Toprim) domain located peripherally. ATP binding at the ATPase domain stabilizes the insertion region within the ATPase domain, keeping the Toprim domain in a closed state. Upon ATP depletion by phages, the Toprim domain opens to bind and cleave the DNA substrate. GajB, which docks on GajA, is activated by the cleaved DNA, ultimately leading to prokaryotic cell death. Our study presents a mechanistic landscape of Gabija activation.
- Department of Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, School of Pharmaceutical Sciences, Wuhan University, Wuhan, China.
Organizational Affiliation: