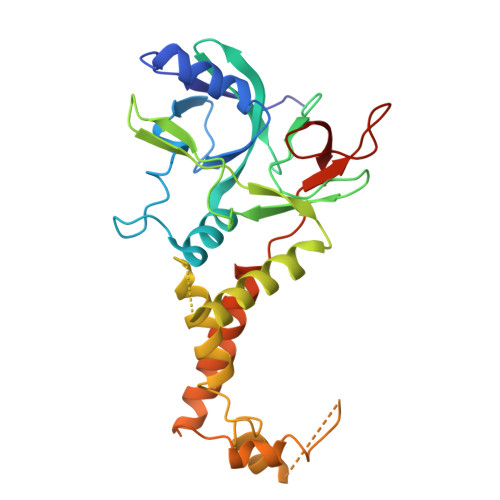
Insights into varicella-zoster virus assembly from the B- and C-capsid at near-atomic resolution structures
Cao, L., Wang, N., Lv, Z., Chen, W., Chen, Z., Song, L., Sha, X., Wang, G., Hu, Y., Lian, X., Cui, G., Fan, J., Quan, Y., Liu, H., Hou, H., Wang, X.(2024) Hlife 2: 64-74