Discovery and mechanistic insights into thieno[3,2-d]pyrimidine and heterocyclic fused pyrimidines inhibitors targeting tubulin for cancer therapy.
Wu, C., Zhang, L., Zhou, Z., Tan, L., Wang, Z., Guo, C., Wang, Y.(2024) Eur J Med Chem 276: 116649-116649
- PubMed: 38972078
- DOI: https://doi.org/10.1016/j.ejmech.2024.116649
- Primary Citation of Related Structures:
8YTX, 8YU9, 8YUA - PubMed Abstract:
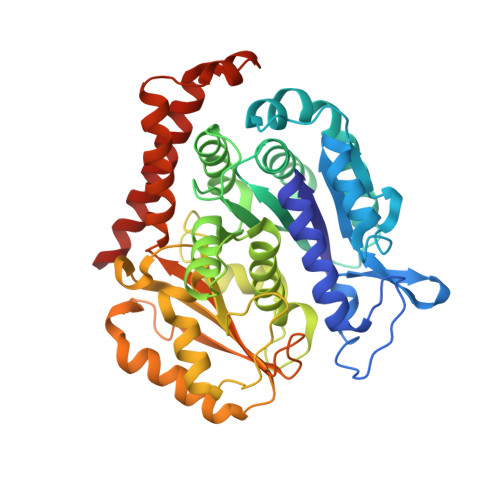
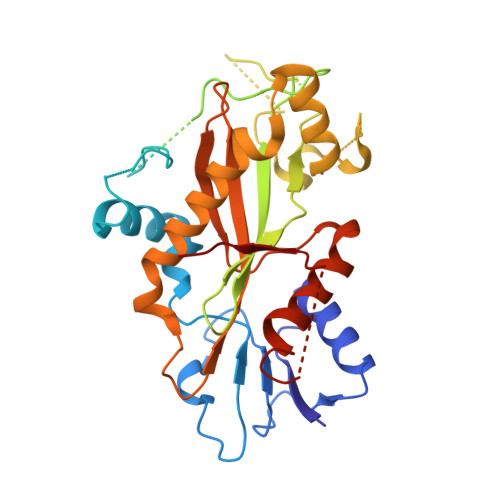
Guided by the X-ray cocrystal structure of the lead compound 4a, we developed a series of thieno[3,2-d]pyrimidine and heterocyclic fused pyrimidines demonstrating potent antiproliferative activity against four tumor cell lines. Two analogs, 13 and 25d, exhibited IC 50 values around 1 nM and overcame P-glycoprotein (P-gp)-mediated multidrug resistance (MDR). At low concentrations, 13 and 25d inhibited both the colony formation of SKOV3 cells in vitro and tubulin polymerization. Furthermore, mechanistic studies showed that 13 and 25d induced G 2 /M phase arrest and apoptosis in SKOV3 cells, as well as dose-dependent inhibition of tumor cell migration and invasion at low concentrations. Most notably, the X-ray cocrystal structures of compounds 4a, 25a, and the optimal molecule 13 in complex with tubulin were elucidated. This study identifies thieno[3,2-d]pyrimidine and heterocyclic fused pyrimidines as representatives of colchicine-binding site inhibitors (CBSIs) with potent antiproliferative activity.
- Department of Pulmonary and Critical Care Medicine, Targeted Tracer Research and Development Laboratory, Institute of Respiratory Health, Frontiers Science Center for Disease-related Molecular Network, National Clinical Research Center for Geriatrics, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, 610041, China.
Organizational Affiliation: