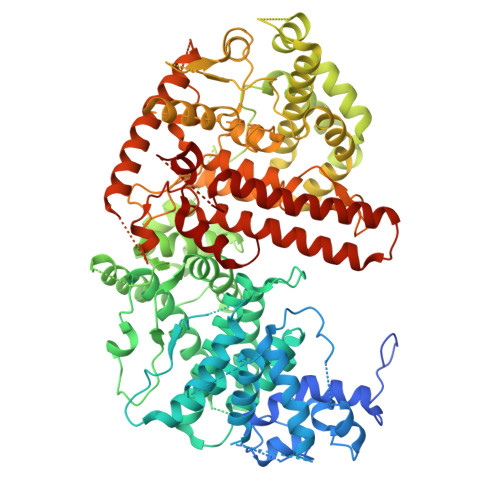
Structural basis of Spliced Leader RNA recognition by the Trypanosoma brucei cap-binding complex.
Bernhard, H., Petrzilkova, H., Popelarova, B., Ziemkiewicz, K., Bartosik, K., Warminski, M., Tengo, L., Groger, H., Dolce, L.G., Mackereth, C.D., Micura, R., Jemielity, J., Kowalinski, E.(2025) Nat Commun 16: 685-685
- PubMed: 39814716
- DOI: https://doi.org/10.1038/s41467-024-55373-w
- Primary Citation of Related Structures:
9F3F, 9F67 - PubMed Abstract:
Kinetoplastids are a clade of eukaryotic protozoans that include human parasitic pathogens like trypanosomes and Leishmania species. In these organisms, protein-coding genes are transcribed as polycistronic pre-mRNAs, which need to be processed by the coupled action of trans-splicing and polyadenylation to yield monogenic mature mRNAs. During trans-splicing, a universal RNA sequence, the spliced leader RNA (SL RNA) mini-exon, is added to the 5'-end of each mRNA. The 5'-end of this mini-exon carries a hypermethylated cap structure and is bound by a trypanosomatid-specific cap-binding complex (CBC). The function of three of the kinetoplastid CBC subunits is unknown, but an essential role in cap-binding and trans-splicing has been suggested. Here, we report cryo-EM structures that reveal the molecular architecture of the Trypanosoma brucei CBC (TbCBC) complex. We find that TbCBC interacts with two distinct features of the SL RNA. The TbCBP20 subunit interacts with the m 7 G cap while TbCBP66 recognizes double-stranded portions of the SL RNA. Our findings pave the way for future research on mRNA maturation in kinetoplastids. Moreover, the observed structural similarities and differences between TbCBC and the mammalian cap-binding complex will be crucial for considering the potential of TbCBC as a target for anti-trypanosomatid drug development.
- EMBL Grenoble, 71 Avenue des Martyrs, Grenoble, France.
Organizational Affiliation: