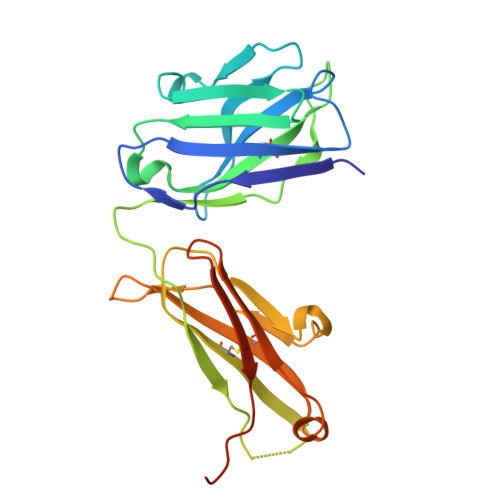
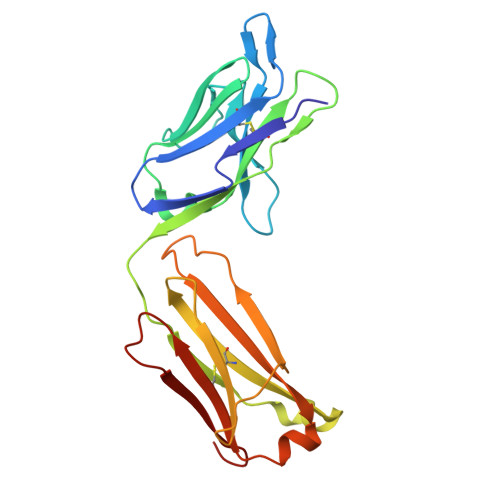
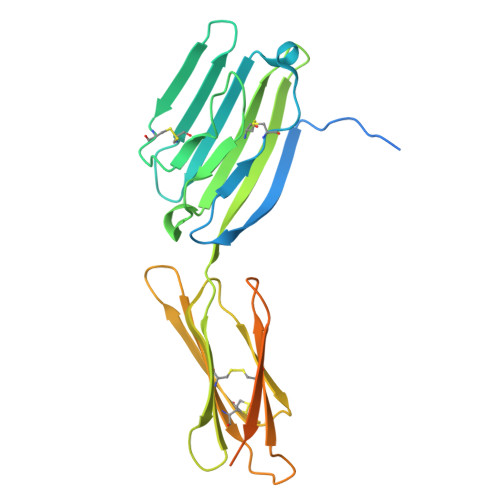
CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody.
Melero, C., Budiardjo, S.J., Daruwalla, A., Larrabee, L., Ganichkin, O., Heiler, A.J., Fenaux, J., Chung, B., Fuh, G., Huang, Y.M.(2024) MAbs 16: 2410316-2410316
- PubMed: 39402718
- DOI: https://doi.org/10.1080/19420862.2024.2410316
- Primary Citation of Related Structures:
9GWT, 9GWZ - PubMed Abstract:
Human CD200R1 (hCD200R1), an immune inhibitory receptor expressed predominantly on T cells and myeloid cells, was identified as a promising immuno-oncology target by the 23andMe database. Blockade of CD200R1-dependent signaling enhances T cell-mediated antitumor activity in vitro and in vivo. 23ME-00610 is a potential first-in-class, humanized IgG1 investigational antibody that binds hCD200R1 with high affinity. We have previously shown that 23ME-00610 inhibits the hCD200R1 immune checkpoint function. Herein, we dissect the molecular mechanism of 23ME-00610 blockade of hCD200R1 by solving the crystal structure of 23ME-00610 Fab in complex with hCD200R1 and performing mutational studies, which show 23ME-00610 blocks the interaction between hCD200 and hCD200R1 through steric hindrance. However, 23ME-00610 does not bind CD200R1 of preclinical species such as cynomolgus monkey MfCD200R1. To enable preclinical toxicology studies of CD200R1 blockade in a pharmacologically relevant non-clinical species, we engineered a surrogate antibody with high affinity toward MfCD200R1. We used phage display libraries of 23ME-00610 variants with individual CDR residues randomized to all 20 amino acids, from which we identified mutations that switched on MfCD200R1 binding. Structural analysis suggests how the surrogate, named 23ME-00611, acquires the ortholog binding ability at the equivalent epitope of 23ME-00610. This engineering approach does not require a priori knowledge of structural and functional mapping of antibody-antigen interaction and thus is generally applicable for therapeutic antibody development when desired ortholog binding is lacking. These findings provide foundational insights as 23ME-00610 advances in clinical studies to gain understanding of the hCD200R1 immune checkpoint as a target in immuno-oncology.
- 23andMe, Therapeutic Unit, South San Francisco, CA, USA.
Organizational Affiliation: