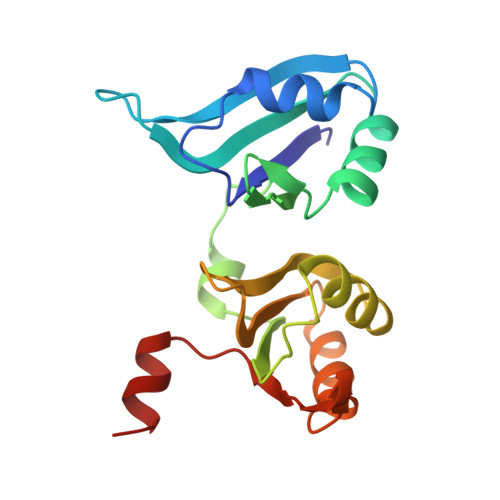
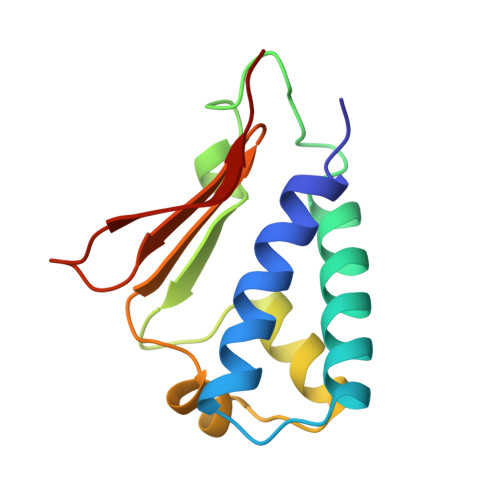
Structural model of the human BTG2-PABPC1 complex by combining mutagenesis, NMR chemical shift perturbation data and molecular docking
Ameerul, A., Almasmoum, H., Pavanello, L., Dominguez, C., Winkler, G.S.(2022) J Mol Biol 434
- PubMed: 35640718
- DOI: https://doi.org/10.1016/j.jmb.2022.167662
- Primary Citation of Related Structures:
9A1R - PubMed Abstract:
Degradation of cytoplasmic mRNA in eukaryotes involves the shortening and removal of the mRNA poly(A) tail by poly(A)-selective ribonuclease (deadenylase) enzymes. In human cells, BTG2 can stimulate deadenylation of poly(A) bound by cytoplasmic poly(A)-binding protein PABPC1. This involves the concurrent binding by BTG2 of PABPC1 and the Caf1/CNOT7 nuclease subunit of the Ccr4-Not deadenylase complex. To understand in molecular detail how PABPC1 and BTG2 interact, we set out to identify amino acid residues of PABPC1 and BTG2 contributing to the interaction. To this end, we first used algorithms to predict PABPC1 interaction surfaces. Comparison of the predicted interaction surface with known residues involved in the binding to poly(A) resulted in the identification of a putative interaction surface for BTG2. Subsequently, we used pulldown assays to confirm the requirement of PABPC1 residues for the interaction with BTG2. Analysis of RNA-binding by PABPC1 variants indicated that PABPC1 residues required for interaction with BTG2 do not interfere with poly(A) binding. After further defining residues of BTG2 that are required for the interaction with PABPC1, we used information from published NMR chemical shift perturbation experiments to guide docking and generate a structural model of the BTG2-PABPC1 complex. A quaternary poly(A)-PABPC1-BTG2-Caf1/CNOT7 model showed that the 3' end of poly(A) RNA is directed towards the catalytic centre of Caf1/CNOT7, thereby providing a rationale for enhanced deadenylation by Caf1/CNOT7 in the presence of BTG2 and PABPC1.
- School of Pharmacy, University of Nottingham, East Drive, University Park, Nottingham NG7 2RD, UK.
Organizational Affiliation: