Molecular architecture and functional dynamics of the pre-incision complex in nucleotide excision repair
Yu, J., Yan, C., Paul, T., Brewer, L., Tsutakawa, S.E., Tsai,C.-L., Hamdan, S.M., Tainer, J.A., Ivanov, I.(2024) Nat Commun
Integrative structure models are generated using different types of input information, including varied experimental data, physical principles, statistical preferences, and other prior information.
Integrative Structure Snapshot
(2024) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription and DNA repair factor IIH helicase subunit XPB | 720 | Homo sapiens | Mutation(s): 0 Gene Names: ERCC3 EC: 5.6.2.4 | 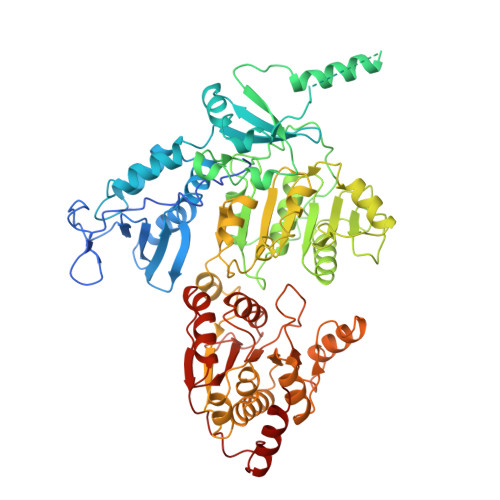 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P19447 (Homo sapiens) Explore P19447 Go to UniProtKB: P19447 | |||||
PHAROS: P19447 GTEx: ENSG00000163161 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19447 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription and DNA repair factor IIH helicase subunit XPD | 760 | Homo sapiens | Mutation(s): 0 Gene Names: ERCC2 EC: 5.6.2.3 | 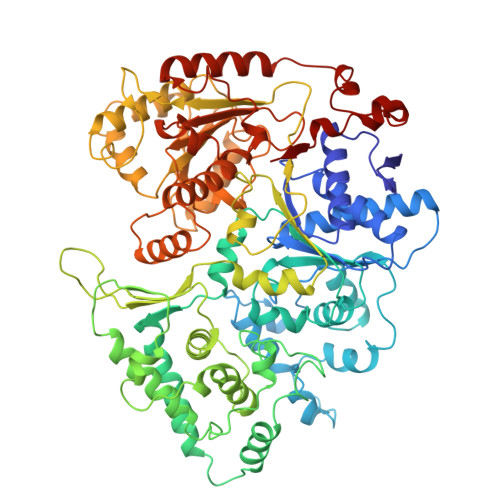 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P18074 (Homo sapiens) Explore P18074 Go to UniProtKB: P18074 | |||||
PHAROS: P18074 GTEx: ENSG00000104884 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P18074 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription factor IIH subunit 4 | 441 | Homo sapiens | Mutation(s): 0 Gene Names: GTF2H4 | 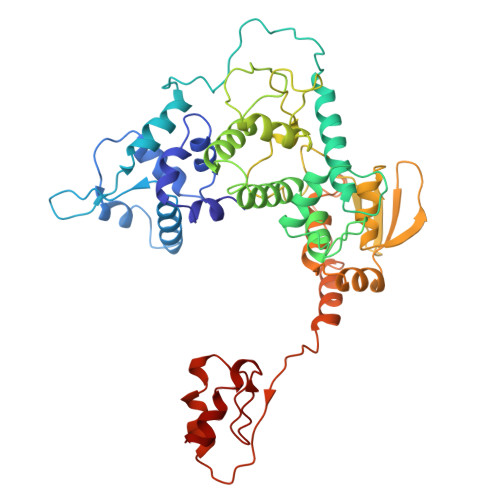 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q92759 (Homo sapiens) Explore Q92759 Go to UniProtKB: Q92759 | |||||
PHAROS: Q92759 GTEx: ENSG00000213780 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92759 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription factor IIH subunit 2 | 377 | Homo sapiens | Mutation(s): 0 Gene Names: GTF2H2 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13888 (Homo sapiens) Explore Q13888 Go to UniProtKB: Q13888 | |||||
GTEx: ENSG00000145736 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13888 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription factor IIH subunit 3 | 292 | Homo sapiens | Mutation(s): 0 Gene Names: GTF2H3 | 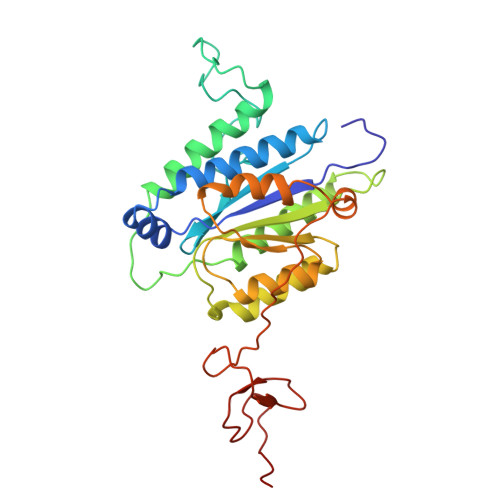 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q13889 (Homo sapiens) Explore Q13889 Go to UniProtKB: Q13889 | |||||
PHAROS: Q13889 GTEx: ENSG00000111358 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q13889 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription factor IIH subunit 5 | 66 | Homo sapiens | Mutation(s): 0 Gene Names: GTF2H5 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q6ZYL4 (Homo sapiens) Explore Q6ZYL4 Go to UniProtKB: Q6ZYL4 | |||||
PHAROS: Q6ZYL4 GTEx: ENSG00000272047 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6ZYL4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA repair protein complementing XP-A cells | 273 | Homo sapiens | Mutation(s): 0 Gene Names: XPA |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P23025 (Homo sapiens) Explore P23025 Go to UniProtKB: P23025 | |||||
PHAROS: P23025 GTEx: ENSG00000136936 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P23025 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| General transcription factor IIH subunit 1 | 154 | Homo sapiens | Mutation(s): 0 Gene Names: GTF2H1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P32780 (Homo sapiens) Explore P32780 Go to UniProtKB: P32780 | |||||
PHAROS: P32780 GTEx: ENSG00000110768 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P32780 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA excision repair protein ERCC-5 | 985 | Homo sapiens | Mutation(s): 0 Gene Names: ERCC5 EC: 3.1 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P28715 (Homo sapiens) Explore P28715 Go to UniProtKB: P28715 | |||||
PHAROS: P28715 GTEx: ENSG00000134899 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28715 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA repair endonuclease XPF Gene: ERCC4, ERCC11, XPF | 227 | Homo sapiens | Mutation(s): 0 Gene Names: ERCC4 EC: 3.1 | 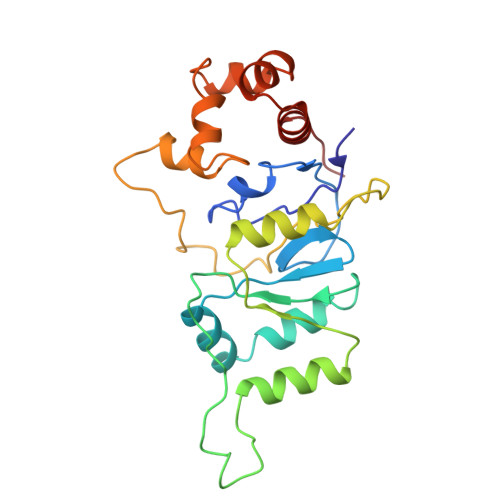 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q92889 (Homo sapiens) Explore Q92889 Go to UniProtKB: Q92889 | |||||
PHAROS: Q92889 GTEx: ENSG00000175595 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q92889 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA excision repair | 198 | Homo sapiens | Mutation(s): 0 Gene Names: ERCC1 | 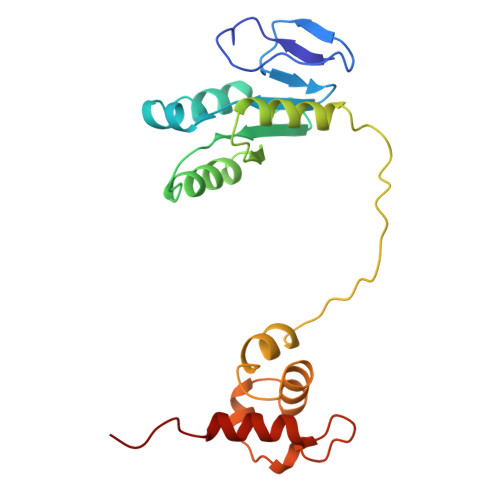 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P07992 (Homo sapiens) Explore P07992 Go to UniProtKB: P07992 | |||||
PHAROS: P07992 GTEx: ENSG00000012061 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07992 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication protein A 70 kDa DNA-binding subunit, N-terminally processed | 434 | Homo sapiens | Mutation(s): 0 Gene Names: RPA1 | 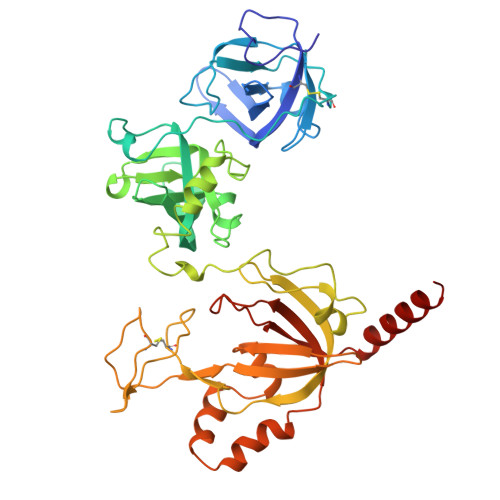 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P27694 (Homo sapiens) Explore P27694 Go to UniProtKB: P27694 | |||||
PHAROS: P27694 GTEx: ENSG00000132383 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P27694 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication protein A 14 kDa subunit | 115 | Homo sapiens | Mutation(s): 0 Gene Names: RPA3 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P35244 (Homo sapiens) Explore P35244 Go to UniProtKB: P35244 | |||||
PHAROS: P35244 GTEx: ENSG00000106399 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P35244 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replication protein A 32 kDa subunit | 225 | Homo sapiens | Mutation(s): 0 Gene Names: RPA2 | 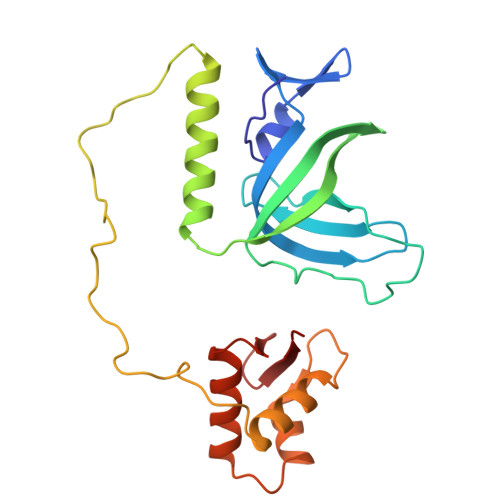 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P15927 (Homo sapiens) Explore P15927 Go to UniProtKB: P15927 | |||||
PHAROS: P15927 GTEx: ENSG00000117748 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P15927 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (66-MER) | O [auth X] | 66 | N/A |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (66-MER) | P [auth Y] | 66 | N/A |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SF4 Query on SF4 | Q [auth B] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | R [auth D] S [auth D] T [auth D] U [auth E] V [auth E] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| MG Query on MG | X [auth J], Z [auth Y] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
Integrative Structure Snapshot