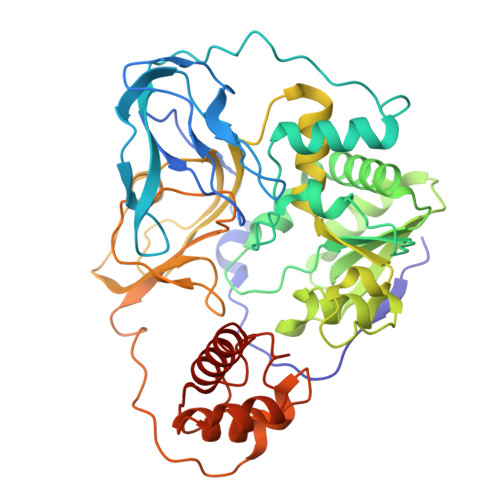
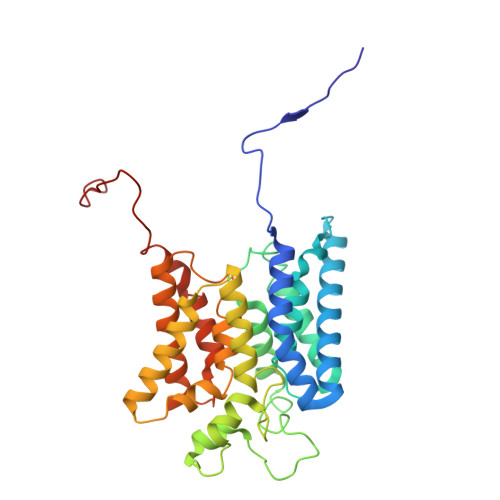
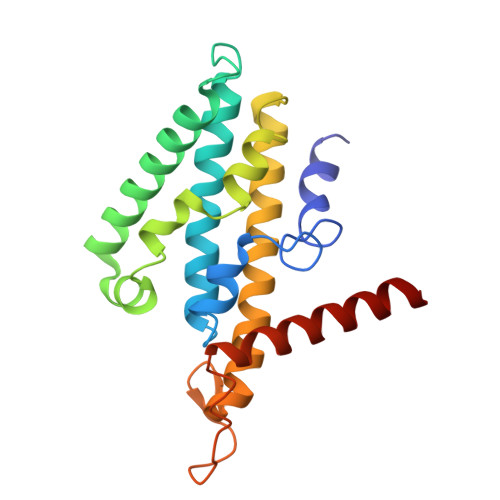
Molecular principles of redox-coupled sodium pumping of the ancient Rnf machinery.
Kumar, A., Roth, J., Kim, H., Saura, P., Bohn, S., Reif-Trauttmansdorff, T., Schubert, A., Kaila, V.R.I., Schuller, J.M., Muller, V.(2025) Nat Commun 16: 2302-2302
- PubMed: 40055346
- DOI: https://doi.org/10.1038/s41467-025-57375-8
- Primary Citation of Related Structures:
9ERI, 9ERJ, 9ERK, 9ERL - PubMed Abstract:
The Rnf complex is the primary respiratory enzyme of several anaerobic prokaryotes that transfers electrons from ferredoxin to NAD + and pumps ions (Na + or H + ) across a membrane, powering ATP synthesis. Rnf is widespread in primordial organisms and the evolutionary predecessor of the Na + -pumping NADH-quinone oxidoreductase (Nqr). By running in reverse, Rnf uses the electrochemical ion gradient to drive ferredoxin reduction with NADH, providing low potential electrons for nitrogenases and CO 2 reductases. Yet, the molecular principles that couple the long-range electron transfer to Na + translocation remain elusive. Here, we resolve key functional states along the electron transfer pathway in the Na + -pumping Rnf complex from Acetobacterium woodii using redox-controlled cryo-electron microscopy that, in combination with biochemical functional assays and atomistic molecular simulations, provide key insight into the redox-driven Na + pumping mechanism. We show that the reduction of the unique membrane-embedded [2Fe2S] cluster electrostatically attracts Na + , and in turn, triggers an inward/outward transition with alternating membrane access driving the Na + pump and the reduction of NAD + . Our study unveils an ancient mechanism for redox-driven ion pumping, and provides key understanding of the fundamental principles governing energy conversion in biological systems.
- SYNMIKRO Research Center and Department of Chemistry, Philipps-University of Marburg, Marburg, Germany.
Organizational Affiliation: