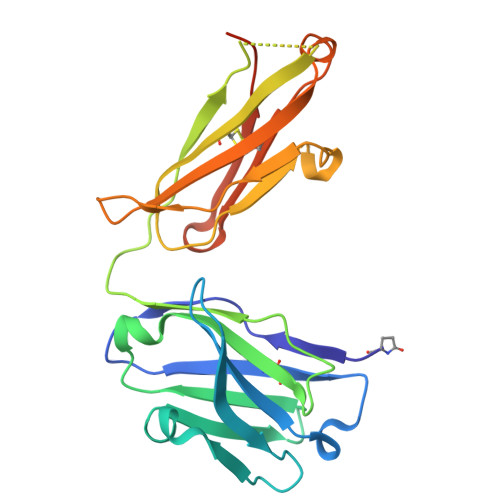
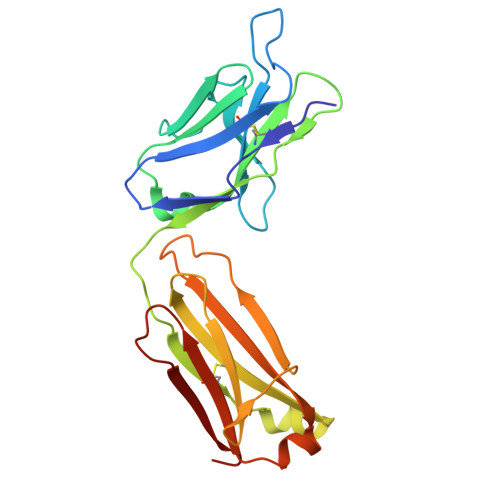
Improved Targeted Delivery of Antisense Oligonucleotide Conjugates with the Antibody Mask
Hung-En, H., Zanini, C., Simonneau, C., Fraidling, J., Kraft, T., Mayer, K., Sommer, A., Indlekofer, A., Wirth, T., Benz, J., Georges, G., Langer, M., Gassner, C., Larraillet, V., Manso, M., Ravn, J., Hofer, K., Emrich, T., Niewoehner, J., Schumacher, F., Brinkmann, U.To be published.