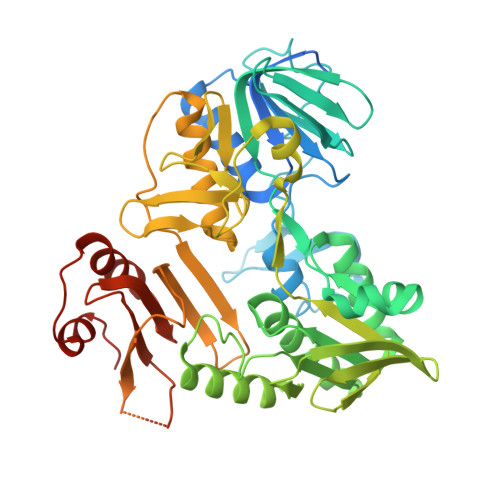
Interaction with AK2A links AIFM1 to cellular energy metabolism.
Rothemann, R.A., Pavlenko, E., Mondal, M., Gerlich, S., Grobushkin, P., Mostert, S., Racho, J., Weiss, K., Stobbe, D., Stillger, K., Lapacz, K., Salscheider, S.L., Petrungaro, C., Ehninger, D., Nguyen, T.H.D., Dengjel, J., Neundorf, I., Bano, D., Poepsel, S., Riemer, J.(2025) Mol Cell 85: 2550
- PubMed: 40578348
- DOI: https://doi.org/10.1016/j.molcel.2025.05.036
- Primary Citation of Related Structures:
9GQY, 9GQZ, 9GR0 - PubMed Abstract:
Apoptosis-inducing factor 1 (AIFM1) is a flavoprotein essential for mitochondrial function and biogenesis. Its interaction with MIA40/CHCHD4, the central component of the mitochondrial disulfide relay, accounts for some, but not all, aspects of AIFM1 function. We provide a high-confidence AIFM1 interactome that elucidates functional partners within the mitochondrial intermembrane space. We found that AIFM1 binding to adenylate kinase 2 (AK2), an essential enzyme that maintains cellular adenine nucleotide pools, depends on the AK2 C-terminal domain. High-resolution cryoelectron microscopy (cryo-EM) and biochemical analyses showed that both MIA40 and AK2A bind the AIFM1 C-terminal β-sheet domain. Their binding enhances NADH oxidoreductase activity by locking an active dimer conformation and, in the case of MIA40, affecting the cofactor-binding site. The AIFM1-AK2A interaction is important during mitochondrial respiration because AIFM1 serves as a recruiting hub within the IMS, regulating mitochondrial bioenergetic output by creating hotspots of metabolic enzymes.
- Redox Metabolism Group, Institute for Biochemistry, University of Cologne, 50674 Cologne, Germany.
Organizational Affiliation: