Structure of avian Pit54 SRCR 1-2 in complex with hemoglobin
Mikkelsen, J.H., Andersen, C.B.F.To be published.
Experimental Data Snapshot
Starting Models: in silico, experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin subunit alpha-A | 142 | Gallus gallus | Mutation(s): 0 | 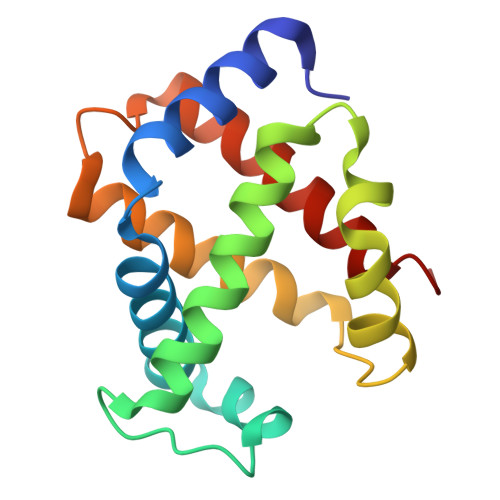 | |
UniProt | |||||
Find proteins for P01994 (Gallus gallus) Explore P01994 Go to UniProtKB: P01994 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01994 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin subunit beta | 147 | Gallus gallus | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P02112 (Gallus gallus) Explore P02112 Go to UniProtKB: P02112 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02112 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PIT54 protein | 508 | Gallus gallus | Mutation(s): 0 Gene Names: PIT54 | 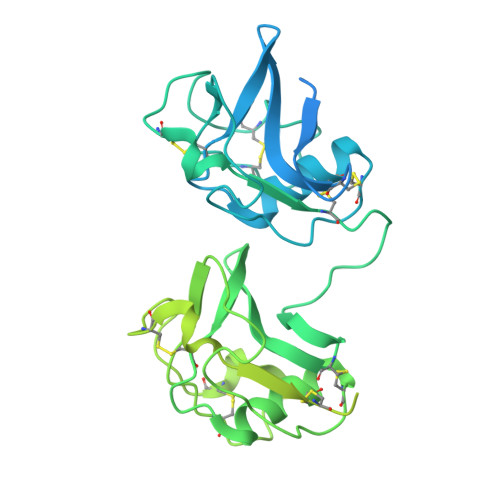 | |
UniProt | |||||
Find proteins for A0A8V0XEL0 (Gallus gallus) Explore A0A8V0XEL0 Go to UniProtKB: A0A8V0XEL0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8V0XEL0 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | M, N | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G32152BH GlyCosmos: G32152BH GlyGen: G32152BH | |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM (Subject of Investigation/LOI) Query on HEM | AA [auth J] P [auth A] Q [auth B] R [auth C] S [auth D] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| CA (Subject of Investigation/LOI) Query on CA | BA [auth K] CA [auth K] DA [auth L] EA [auth L] T [auth E] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 101.534 | α = 90 |
| b = 157.592 | β = 90 |
| c = 175.689 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |