A cooperative binding mechanism steers the recruitment of peripheral subunits in the E. coli pyruvate dehydrogenase complex
Bothe, S.N., Racunica, D., Zajec Hudnik, T., Zdanowicz, R., Bothe, A., Giese, C., Glockshuber, R.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | 66 | Escherichia coli | Mutation(s): 0 Gene Names: aceF, b0115, JW0111 EC: 2.3.1.12 | 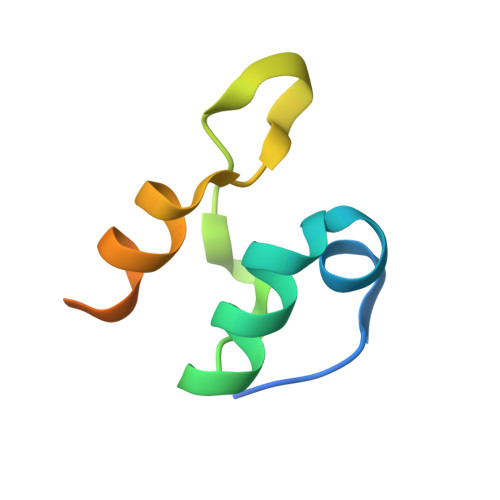 | |
UniProt | |||||
Find proteins for P06959 (Escherichia coli (strain K12)) Explore P06959 Go to UniProtKB: P06959 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06959 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dihydrolipoyl dehydrogenase | 473 | Escherichia coli | Mutation(s): 0 Gene Names: lpdA, lpd, b0116, JW0112 EC: 1.8.1.4 | 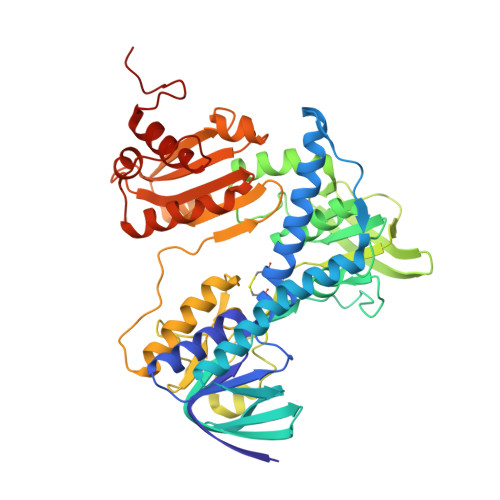 | |
UniProt | |||||
Find proteins for P0A9P0 (Escherichia coli (strain K12)) Explore P0A9P0 Go to UniProtKB: P0A9P0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A9P0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD (Subject of Investigation/LOI) Query on FAD | D [auth B], E [auth C] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 60.703 | α = 90 |
| b = 88.353 | β = 90 |
| c = 200.687 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swiss National Science Foundation | Switzerland | -- |