Crystal structure of the diheme 4D2 (mutant T19D) with bound Fe(III) mesoporphyrin IX
Mellor, C.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4D2 (mutant T19D) | 112 | synthetic construct | Mutation(s): 0 | 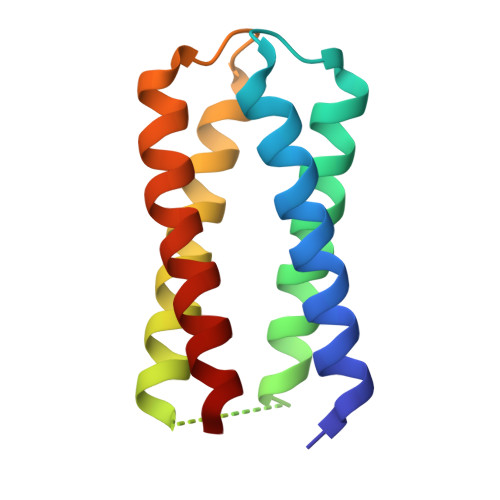 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MH0 (Subject of Investigation/LOI) Query on MH0 | B [auth A], C [auth A] | Mesoheme C34 H36 Fe N4 O4 CKMCSMAXNUVLQI-RGGAHWMASA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 81.187 | α = 90 |
| b = 81.187 | β = 90 |
| c = 58.817 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| xia2.multiplex | data reduction |
| xia2.multiplex | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/W003449/1 |