Collagen VI microfibril structure reveals mechanism for molecular assembly and clustering of inherited pathogenic mutations
Godwin, A., Snee, M., Becker, M., Dajani, R., Collins, R., Roseman, A., Baldock, C.(2025) Nat Commun
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2025) Nat Commun
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Collagen type VI alpha 3 chain | A [auth C], D [auth F] | 3,061 | Bos taurus | Mutation(s): 0 | 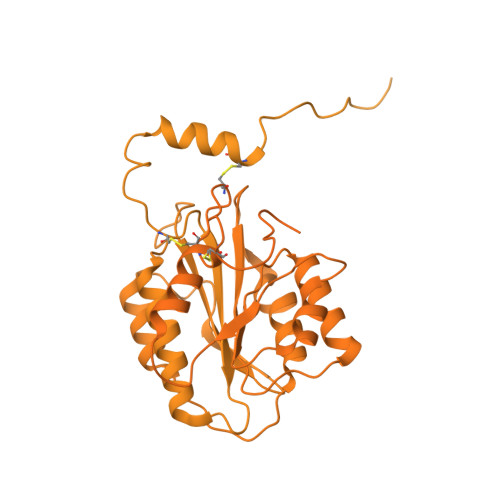 |
UniProt | |||||
Find proteins for A0AAA9TAB4 (Bos taurus) Explore A0AAA9TAB4 Go to UniProtKB: A0AAA9TAB4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0AAA9TAB4 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Collagen type VI alpha 1 chain | B [auth A], E [auth D] | 1,027 | Bos taurus | Mutation(s): 0 | 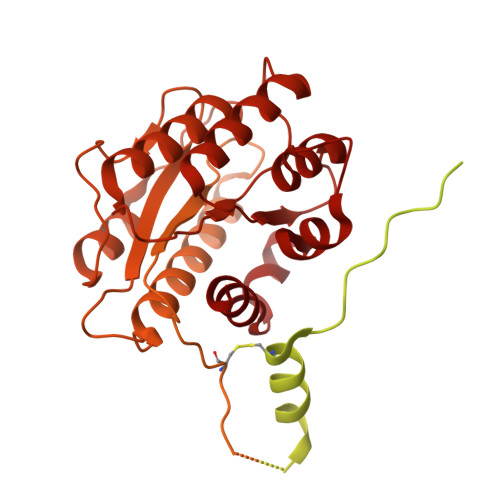 |
UniProt | |||||
Find proteins for E1BI98 (Bos taurus) Explore E1BI98 Go to UniProtKB: E1BI98 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E1BI98 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Collagen type VI alpha 2 chain | C [auth B], F [auth E] | 1,027 | Bos taurus | Mutation(s): 0 | 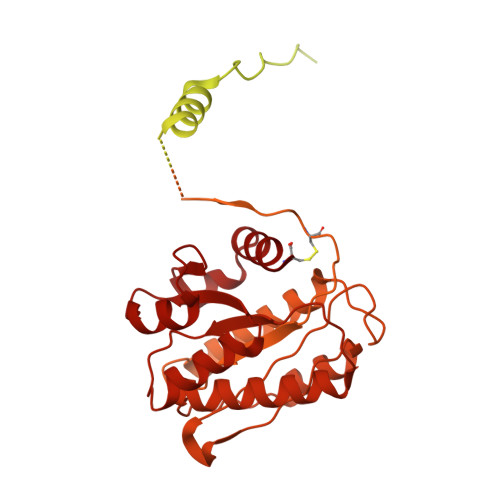 |
UniProt | |||||
Find proteins for A0AAA9TXG2 (Bos taurus) Explore A0AAA9TXG2 Go to UniProtKB: A0AAA9TXG2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0AAA9TXG2 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | M [auth C], N [auth F] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | -- |