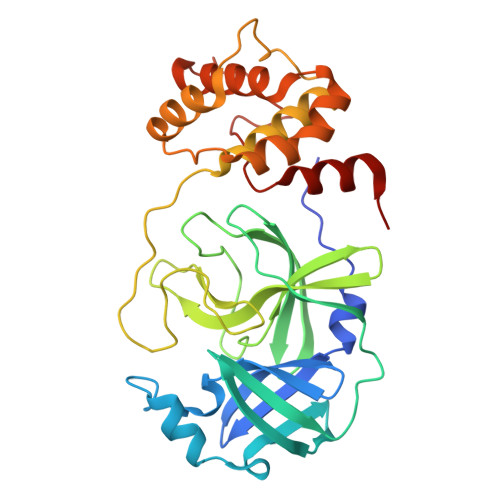
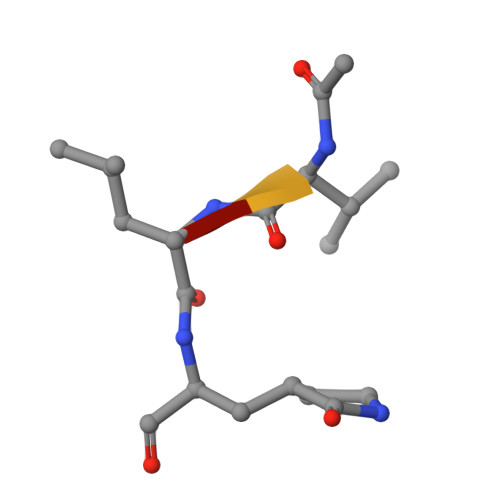
Discovery of an Orally Bioavailable Reversible Covalent SARS-CoV-2 M pro Inhibitor with Pan-Coronavirus Activity.
Tan, Q.W., Vankadara, S., Fong, J.Y., See, Y.Y., Baburajendran, N., Ng, P.S., Xu, W., Yeo, Y.K., Wang, W., Low, C.H., Tan, L.H., Ju Tay, E.G., Wong, Y.X., Huang, C., Sim, S., Ang, S.H., Min Toh, H.H., Mohammad, J., Wang, G., Liu, B., Tan, S.T., Kwek, P.Z., Dawson, M.D., Oh, Q.Y., Koh, X., Joy, J., Lee, M.A., Stunkel, W., Pendharkar, V., Hentze, H., Lim, S.P., Ethirajulu, K., Brian Chia, C.S., Cherian, J.(2025) J Med Chem
- PubMed: 40773370
- DOI: https://doi.org/10.1021/acs.jmedchem.5c00581
- Primary Citation of Related Structures:
9KGJ, 9KGN, 9KGQ, 9KGR, 9KGS - PubMed Abstract:
Resulting in several million deaths globally, the COVID-19 pandemic has highlighted the criticality of antiviral drugs during a viral pandemic. Herein, we describe our efforts toward targeting SARS-CoV-2 M pro , a key viral protease, which led to the discovery of compound 18 , a reversible covalent inhibitor with potent antiviral activity against several clinical variants of SARS-CoV-2. Compound 18 demonstrated dose-dependent efficacy in a mouse-adapted SARS-CoV-2 infection model, with favorable pharmacokinetic profiles in mice, rats, dogs, and monkeys.
Organizational Affiliation:
Experimental Drug Development Centre (EDDC), Agency for Science, Technology and Research (A*STAR), 10 Biopolis Road, Chromos #05-01, Singapore 138670, Republic of Singapore.