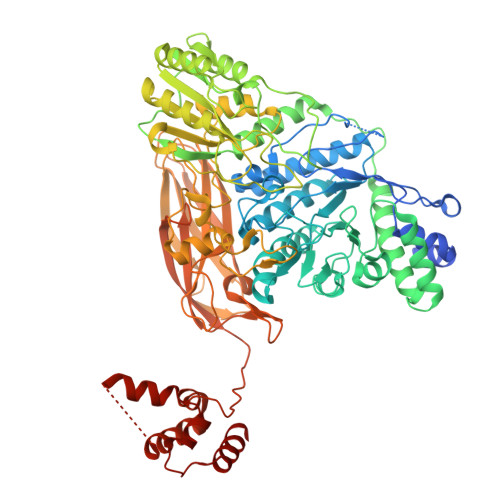
Structural and functional implications of the novel C-terminal domain and flexible loop in unique beta-glucosidase hydrolyzing sesaminol triglucoside to produce sesaminol
Yanai, T., Arai, A., Takahashi, Y., Imaizumi, R., Takeshita, K., Matsuura, H., Sakai, N., Takahashi, S., Yamamoto, M., Kataoka, K., Nakayama, T., Yamashita, S.To be published.