An electron-bifurcating "plug" to a protein nanowire in tungsten-dependent aldehyde detoxification.
Feng, X., Schut, G.J., Putumbaka, S., Li, H., Adams, M.W.W.(2025) Proc Natl Acad Sci U S A 122: e2501900122-e2501900122
- PubMed: 40694326
- DOI: https://doi.org/10.1073/pnas.2501900122
- Primary Citation of Related Structures:
9MQX - PubMed Abstract:
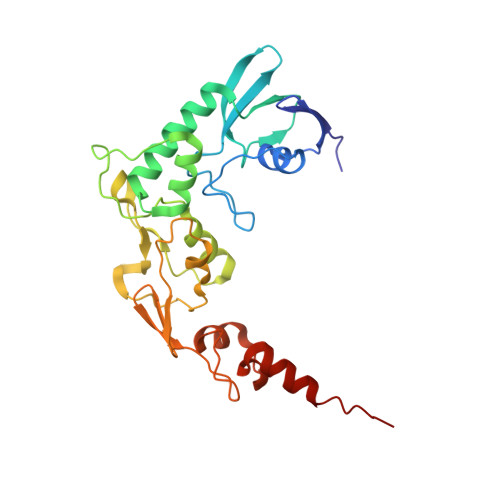
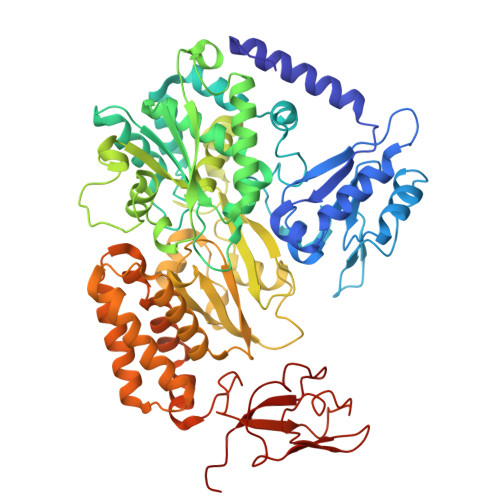
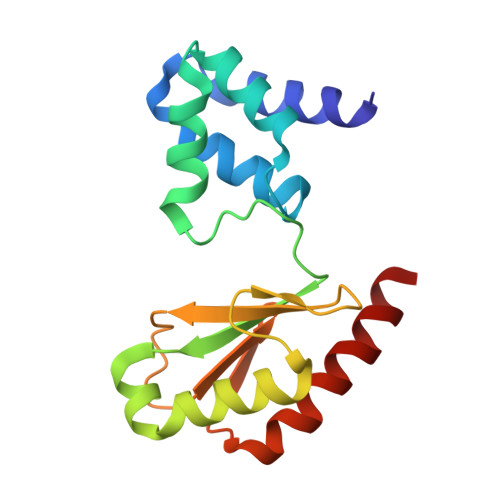
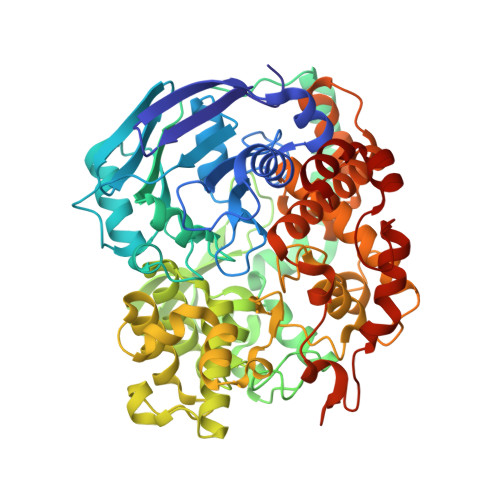
Members of the tungsten-containing oxidoreductase (WOR) family, which contain a tungstopyranopterin (Tuco) cofactor, are typically either monomeric (WorL) or heterodimeric (WorLS). These enzymes oxidize aldehydes to the corresponding acids while reducing the redox protein ferredoxin. They have been structurally characterized mainly using WORs from hyperthermophilic archaea. The WORs of some bacteria contain three additional subunits of the BfuABC family and these chimeric WorABCSL enzymes catalyze an electron-bifurcating reaction in which aldehyde oxidation is coupled to the simultaneous reduction of ferredoxin and nicotinamide adenine dinucleotide. In human gut microbes, electron bifurcation by WorABSL is proposed to enable the detoxification of aldehydes generated from cooked foods and in the tungstocentric production of beneficial short chain fatty acids from lactate, potentially impacting health. Herein we present the high-resolution cryogenic electron microscopy (cryo-EM) structure of the WorABCSL purified from the bacterium Acetomicrobium mobile. The structure reveals a surprising 1:3 stoichiometry between WorABC and WorSL, with the WorSL units forming a nanowire-like architecture leading from three Tuco-containing catalytic sites in WorL via strings of multiple iron-sulfur clusters in WorS to a single bifurcating WorABC core. Our structure uncovers a distinct domain arrangement that links three Tuco-dependent aldehyde oxidation sites with the bifurcation process and potentially facilitates environmental aldehyde oxidation.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503.
Organizational Affiliation: