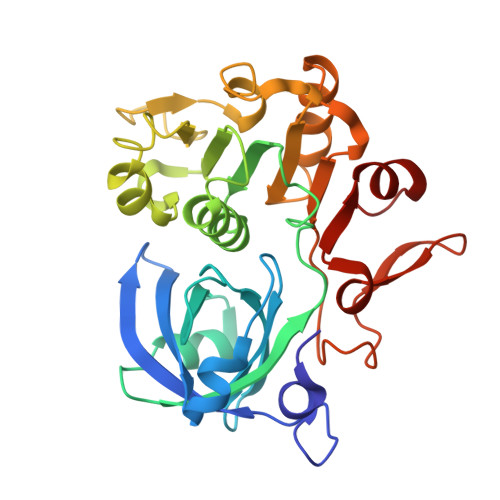
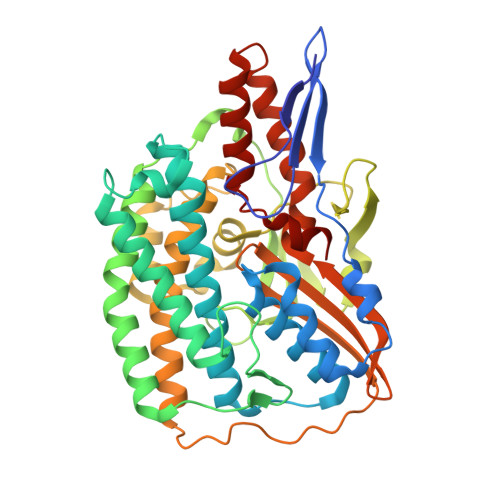
Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Xiao, X., Schut, G.J., Feng, X., McTernan, P.M., Haja, D.K., Lanzilotta, W.N., Adams, M.W.W., Li, H.(2025) Structure 33: 1470
- PubMed: 40570843
- DOI: https://doi.org/10.1016/j.str.2025.05.017
- Primary Citation of Related Structures:
9E15, 9E1J, 9NEZ, 9NF0 - PubMed Abstract:
The cytoplasmic hydrogenase I (SHI) from the hyperthermophilic archaeon Pyrococcus furiosus belongs to the group III hydrogenase family. SHI oxidizes NADPH rather than NADH to reduce protons and evolve hydrogen gas, and because of this property, coupled with its high thermal stability, the enzyme holds great potential for economical hydrogen production. Despite decades of efforts, the SHI structure has remained unknown. Here, we report the cryoelectron microscopic (cryo-EM) structures of the heterotetrameric SHI holoenzyme (αδβγ). SHI is a symmetric dimer of two individually functional heterotetramers. SHI-αδ resembles the standard [NiFe] hydrogenase, and SHI-βγ function as the NADPH oxidoreductase. SHI-β contains three [4Fe-4S] clusters that relay electrons from NADPH in SHI-γ to the catalytic [NiFe] cluster in SHI-αδ for H 2 production. These structures will guide the adaptation of this unique enzyme for biotechnological applications.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503, USA.
Organizational Affiliation: