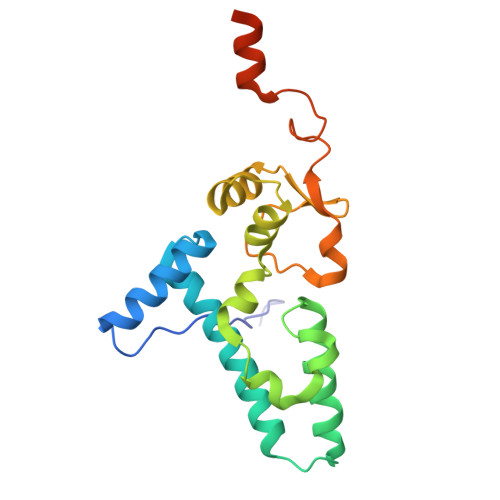
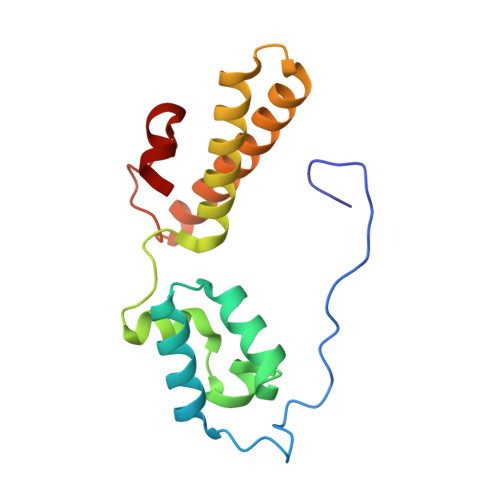
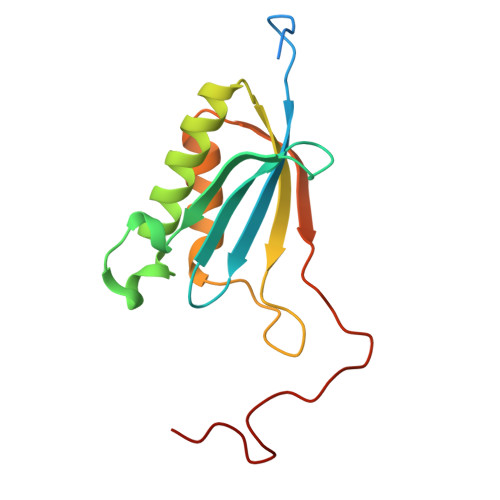
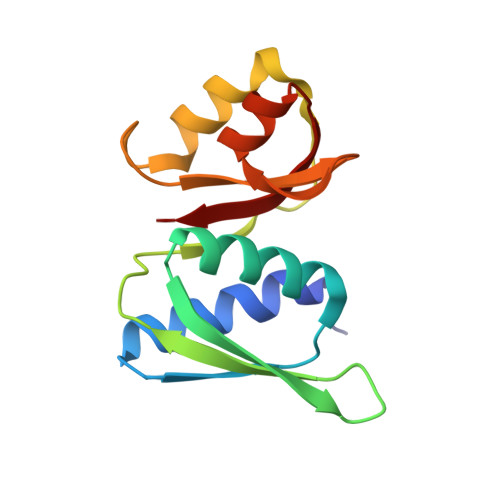
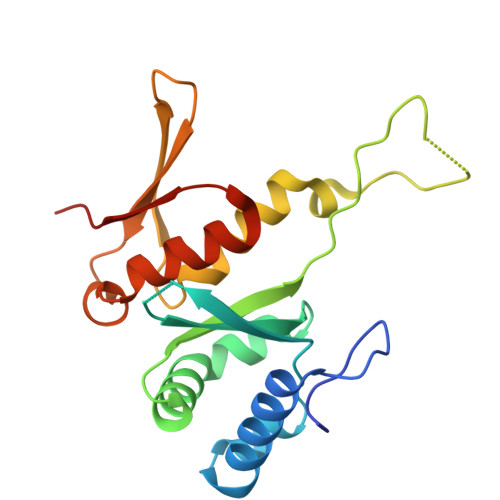
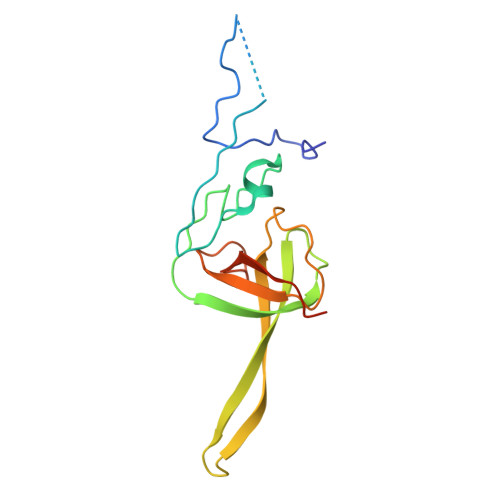
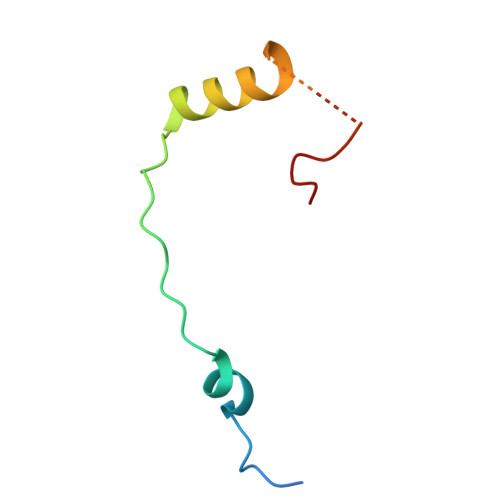
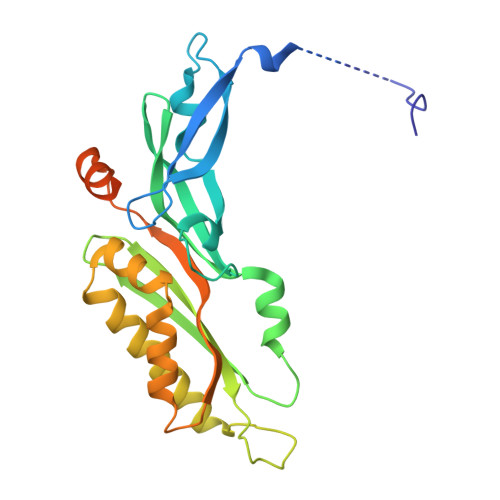
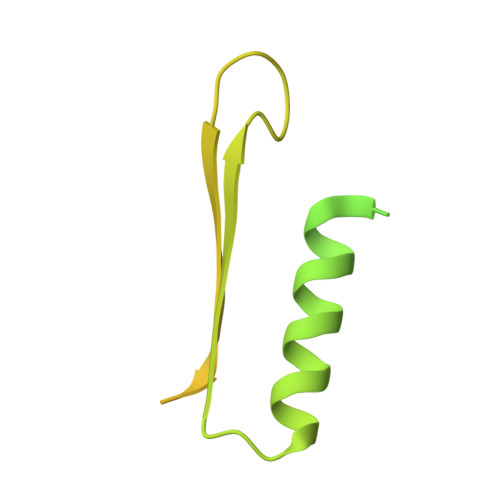
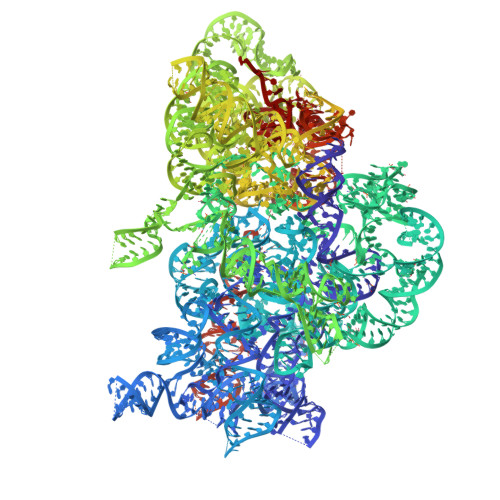
SARS-CoV-2 nsp1 mediates broad inhibition of translation in mammals.
Gen, R., Addetia, A., Asarnow, D., Park, Y.J., Quispe, J., Chan, M.C., Brown, J.T., Lee, J., Campbell, M.G., Lapointe, C.P., Veesler, D.(2025) Cell Rep 44: 115696-115696
- PubMed: 40359110
- DOI: https://doi.org/10.1016/j.celrep.2025.115696
- Primary Citation of Related Structures:
9NPX, 9NPY - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) non-structural protein 1 (nsp1) promotes innate immune evasion by inhibiting host translation in human cells. However, the role of nsp1 in other host species remains elusive, especially in bats-natural reservoirs of sarbecoviruses with a markedly different innate immune system than humans. We reveal that nsp1 potently inhibits translation in Rhinolophus lepidus bat cells, which belong to the same genus as known sarbecovirus reservoir hosts. We determined a cryoelectron microscopy structure of nsp1 bound to the R. lepidus 40S ribosomal subunit, showing that it blocks the mRNA entry channel by targeting a highly conserved site among mammals. Accordingly, we found that nsp1 blocked protein translation in mammalian cells from several species, underscoring its broadly inhibitory activity and conserved role in numerous SARS-CoV-2 hosts. Our findings illuminate the arms race between coronaviruses and mammalian host immunity, providing a foundation for understanding the determinants of viral maintenance in bat hosts and spillover.
- Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: