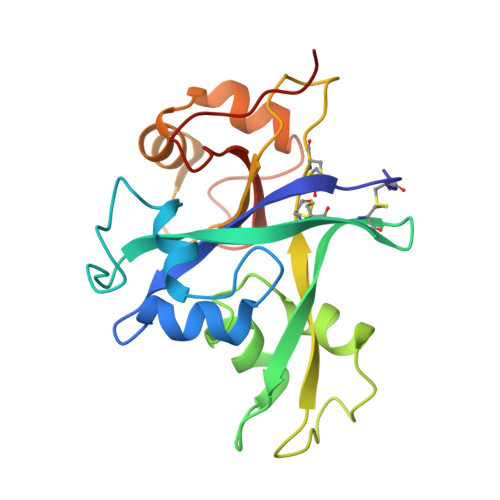
Structural basis for size-selective perception of chitin in plants
Gysel, K., Hansen, S.B., Ruebsam, H., Alsarraf, H.M.A.B., Madland, E., Cheng, J.X.J., Baadegaard, C., Poulsen, E.C., Vinther, M., Fort, S., Stougaard, J., Andersen, K.R.To be published.