Substrate specificity and mechanism of action of the HerA-NurA nuclease from the hyperthermophilic archaeon Thermococcus kodakarensis
Uda, K., Numata, T.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA helicase | 592 | Thermococcus kodakarensis | Mutation(s): 0 Gene Names: TK2213 EC: 3.6.4.12 | 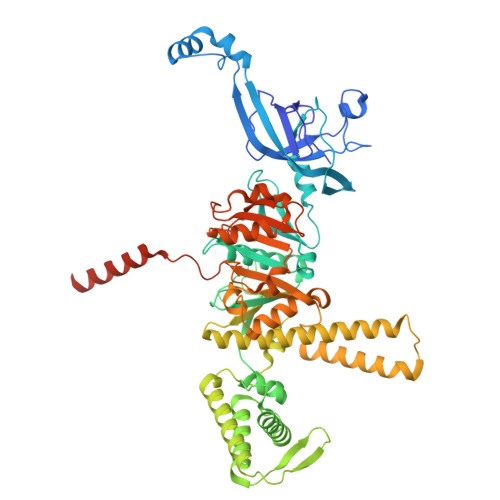 | |
UniProt | |||||
Find proteins for Q5JHP7 (Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1)) Explore Q5JHP7 Go to UniProtKB: Q5JHP7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5JHP7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-3' nuclease, encoded next to Rad50 and Mre11 homologs | 443 | Thermococcus kodakarensis | Mutation(s): 1 Gene Names: TK2210 | 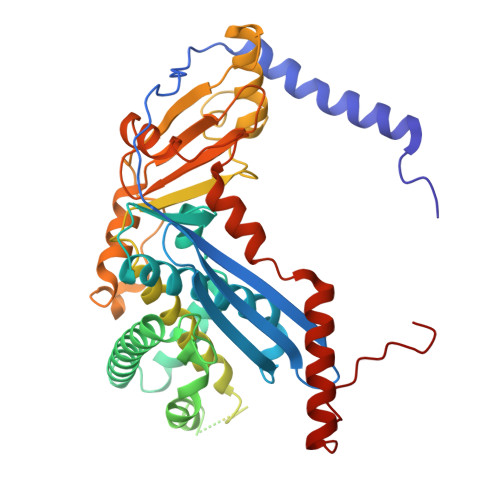 | |
UniProt | |||||
Find proteins for Q5JHL5 (Thermococcus kodakarensis (strain ATCC BAA-918 / JCM 12380 / KOD1)) Explore Q5JHL5 Go to UniProtKB: Q5JHL5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q5JHL5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA | 13 | DNA molecule | 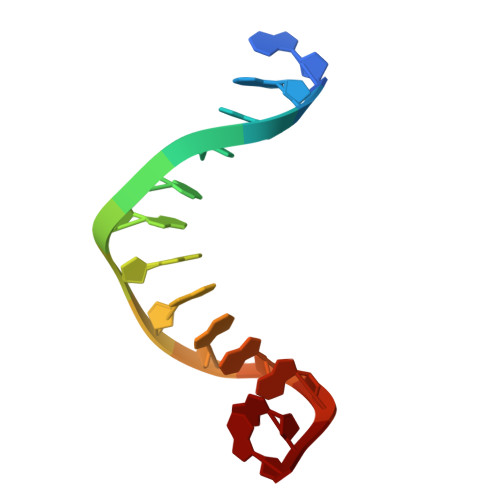 | ||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA | 13 | DNA molecule | 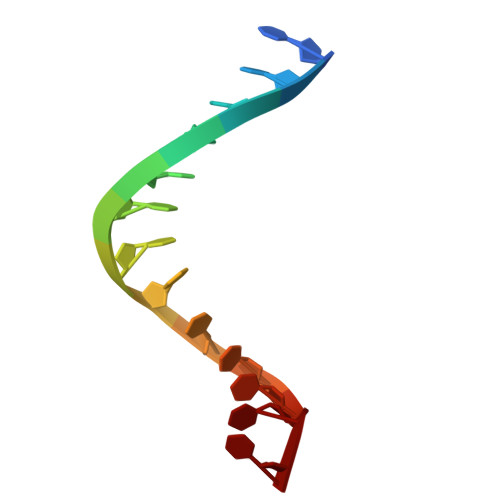 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AGS (Subject of Investigation/LOI) Query on AGS | K [auth A], M [auth B], T [auth F] | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER C10 H16 N5 O12 P3 S NLTUCYMLOPLUHL-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | O [auth C], P [auth D], R [auth E] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MN (Subject of Investigation/LOI) Query on MN | L [auth A], N [auth B], Q [auth D], S [auth E], U [auth F] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | |
| MODEL REFINEMENT | PHENIX | 1.19.2_4158 |
| MODEL REFINEMENT | Coot |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | JP19K22289 |
| Japan Society for the Promotion of Science (JSPS) | Japan | JP21K05394 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 20H02916 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 20K21281 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 24H00505 |