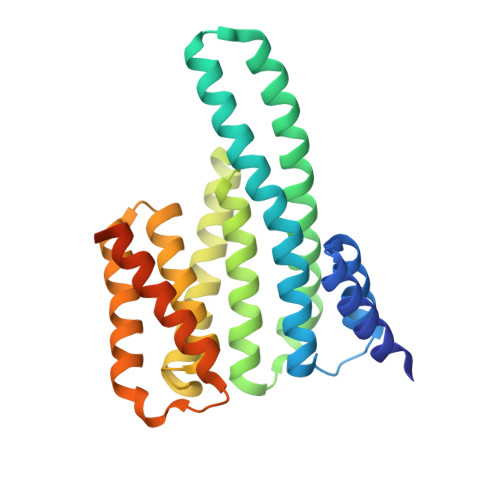
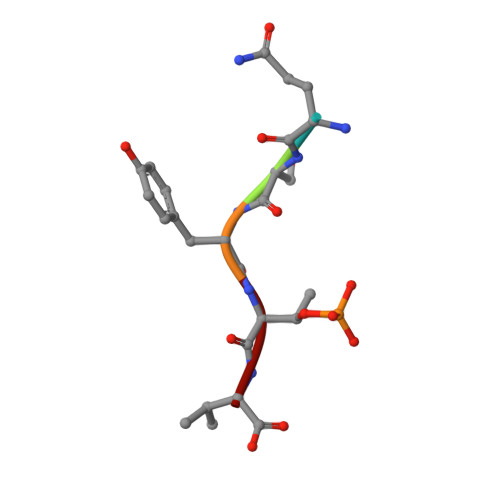
A structural rationale for selective stabilization of anti-tumor interactions of 14-3-3 proteins by cotylenin A
Ottmann, C., Weyand, M., Sassa, T., Inoue, T., Kato, N., Wittinghofer, A., Oecking, C.(2009) J Mol Biology 386: 913-919
- PubMed: 19244612
- DOI: https://doi.org/10.1016/j.jmb.2009.01.005
- Primary Citation of Related Structures:
3E6Y - PubMed Abstract:
Cotylenin A, a fungal metabolite originally described as a cytokinin-like bioactive substance against plants shows differentiation-inducing and anti-tumor activity in certain human cancers. Here, we present the crystal structure of cotylenin A acting on a 14-3-3 regulatory protein complex. By comparison with the closely related, but non-anticancer agent fusicoccin A, a rationale for the activity of cotylenin A in human cancers is presented. This class of fusicoccane diterpenoids are possible general modulators of 14-3-3 protein-protein interactions. In this regard, specificities for individual 14-3-3/target protein complexes might be achieved by varying the substituent pattern of the diterpene ring system. As the different activities of fusicoccin A and cotylenin A in human cancers suggest, hydroxylation of C12 might be a sufficient determinant of structural specificity.
- Chemical Genomics Centre, Max Planck Society, Dortmund, Germany. christian.ottmann@cgc.mpg.de
Organizational Affiliation: