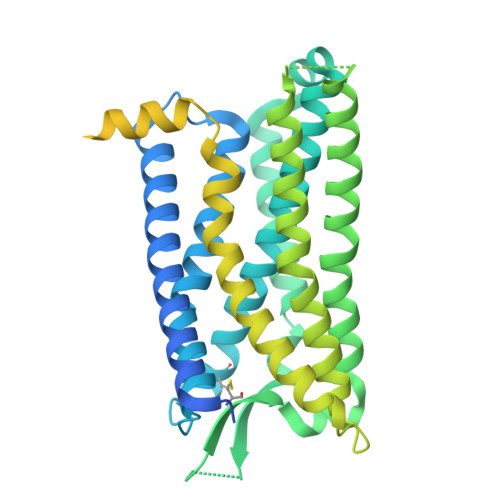
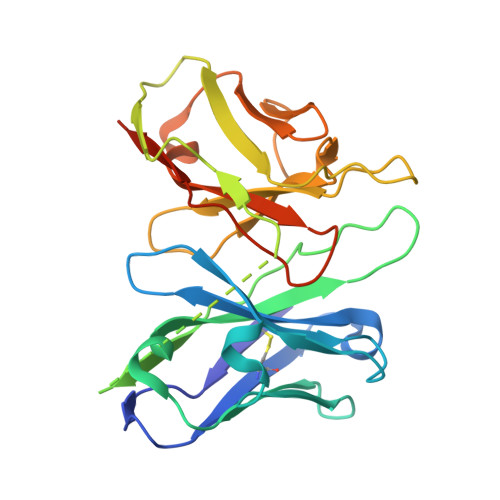
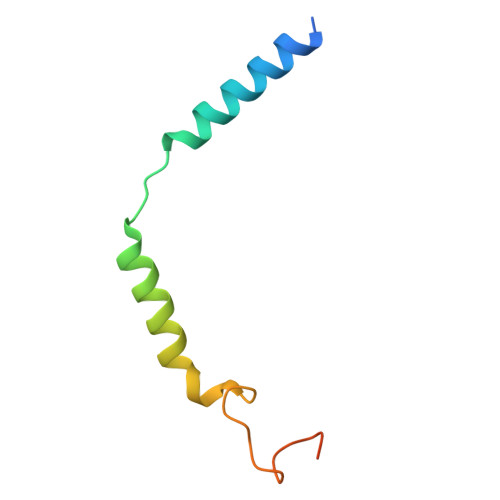
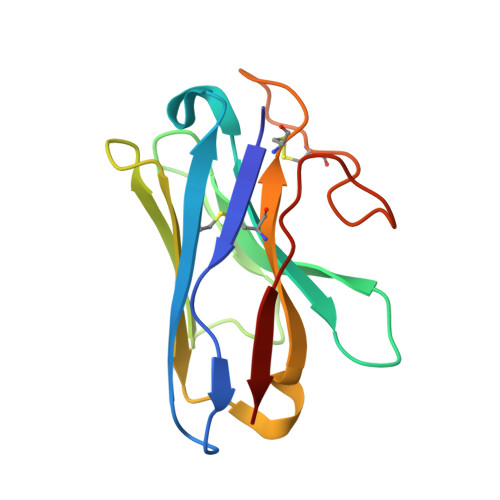
The structure and function of the ghrelin receptor coding for drug actions.
Shiimura, Y., Im, D., Tany, R., Asada, H., Kise, R., Kurumiya, E., Wakasugi-Masuho, H., Yasuda, S., Matsui, K., Kishikawa, J.I., Kato, T., Murata, T., Kojima, M., Iwata, S., Masuho, I.(2025) Nat Struct Mol Biol 32: 531-542
- PubMed: 39833471
- DOI: https://doi.org/10.1038/s41594-024-01481-6
- Primary Citation of Related Structures:
8JSR - PubMed Abstract:
Drugs targeting the ghrelin receptor hold therapeutic potential in anorexia, obesity and diabetes. However, developing effective drugs is challenging. To tackle this common issue across a broad drug target, this study aims to understand how anamorelin, the only approved drug targeting the ghrelin receptor, operates compared to other synthetic drugs. Our research elucidated the receptor's structure with anamorelin and miniG q , unveiling anamorelin's superagonistic activity. We demonstrated that ligands with distinct chemical structures uniquely bind to the receptor, resulting in diverse conformations and biasing signal transduction. Moreover, our study showcased the utility of structural information in effectively identifying natural genetic variations altering drug action and causing severe functional deficiencies, offering a basis for selecting the right medication on the basis of the individual's genomic sequence. Thus, by building on structural analysis, this study enhances the foundational framework for selecting therapeutic agents targeting the ghrelin receptor, by effectively leveraging signaling bias and genetic variations.
- Division of Molecular Genetics, Institute of Life Science, Kurume University, Fukuoka, Japan. shiimura_yuuki@kurume-u.ac.jp.
Organizational Affiliation: