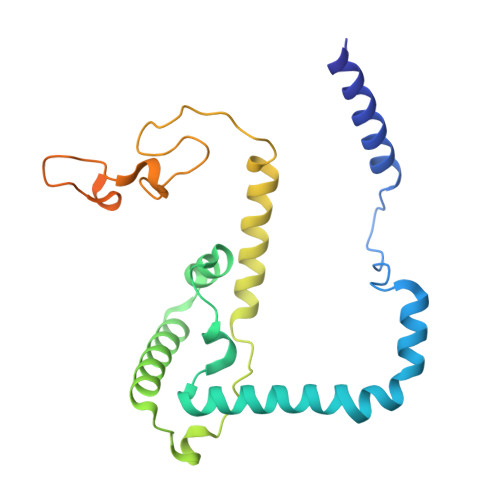
Structure, assembly and inhibition of the Toxoplasma gondii respiratory chain supercomplex.
MacLean, A.E., Shikha, S., Ferreira Silva, M., Gramelspacher, M.J., Nilsen, A., Liebman, K.M., Pou, S., Winter, R.W., Meir, A., Riscoe, M.K., Doggett, J.S., Sheiner, L., Muhleip, A.(2025) Nat Struct Mol Biol 32: 1424-1433
- PubMed: 40389671
- DOI: https://doi.org/10.1038/s41594-025-01531-7
- Primary Citation of Related Structures:
9G9T, 9H8T, 9I4X - PubMed Abstract:
The apicomplexan mitochondrial electron transport chain is essential for parasite survival and displays a divergent subunit composition. Here we report cryo-electron microscopy structures of an apicomplexan III 2 -IV supercomplex and of the drug target complex III 2 . The supercomplex structure reveals how clade-specific subunits form an apicomplexan-conserved III 2 -IV interface with a unique, kinked architecture, suggesting that supercomplexes evolved independently in different eukaryotic lineages. A knockout resulting in supercomplex disassembly challenges the proposed role of III 2 -IV in electron transfer efficiency as suggested for mammals. Nevertheless, knockout analysis indicates that III 2 -IV is critical for parasite fitness. The complexes from the model parasite Toxoplasma gondii were inhibited with the antimalarial atovaquone, revealing interactions underpinning species specificity. They were also inhibited with endochin-like quinolone (ELQ)-300, an inhibitor in late-stage preclinical development. Notably, in the apicomplexan binding site, ELQ-300 is flipped compared with related compounds in the mammalian enzyme. On the basis of the binding modes and parasite-specific interactions discovered, we designed more potent ELQs with subnanomolar activity against T. gondii. Our findings reveal critical evolutionary differences in the role of supercomplexes in mitochondrial biology and provide insight into cytochrome b inhibition, informing future drug discovery.
Organizational Affiliation:
School of Infection and Immunity, University of Glasgow, Glasgow, UK.