CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
Kerff, F., Fonze, E., Sauvage, E., Frere, J.M., Charlier, P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-lactamase PSE-2 | 248 | Pseudomonas aeruginosa | Mutation(s): 0 Gene Names: plasmid pMON234 EC: 3.5.2.6 | 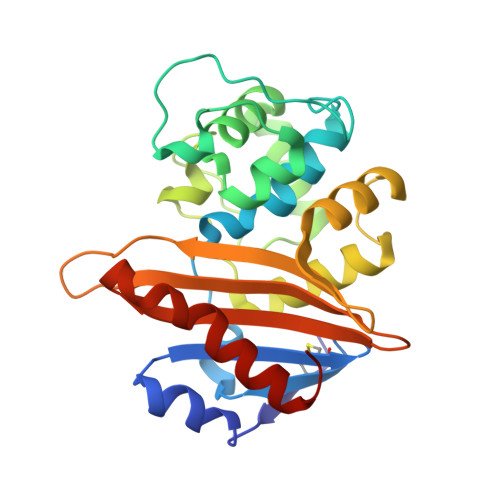 | |
UniProt | |||||
Find proteins for P14489 (Pseudomonas aeruginosa) Explore P14489 Go to UniProtKB: P14489 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14489 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MX1 Query on MX1 | C [auth A], D [auth B] | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO
XYLIC ACID C18 H18 N2 O10 ZTOQXKUYWZBJLG-XNAIMREJSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.706 | α = 90 |
| b = 96.003 | β = 90 |
| c = 125.195 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| AMoRE | phasing |
| CNS | refinement |
| CCP4 | data scaling |