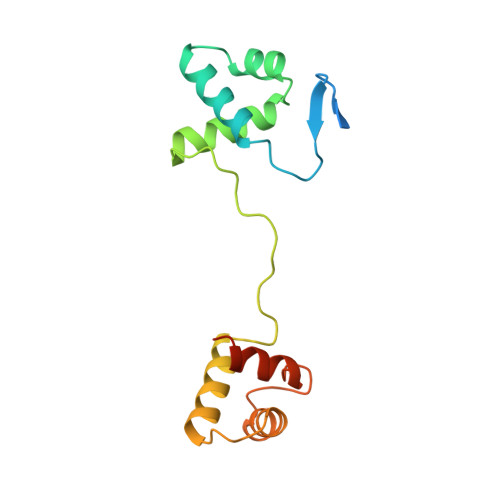
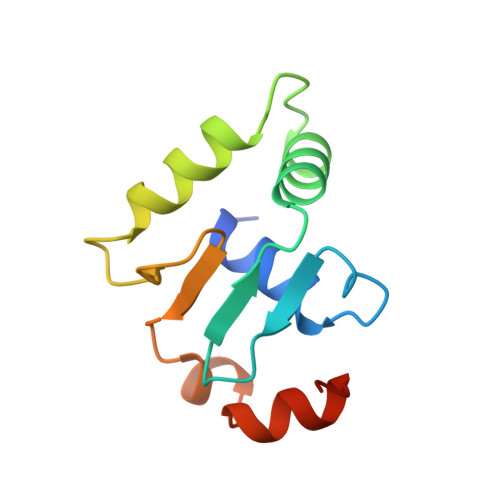
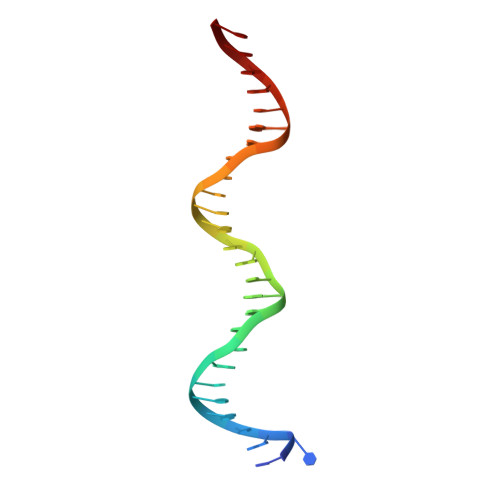
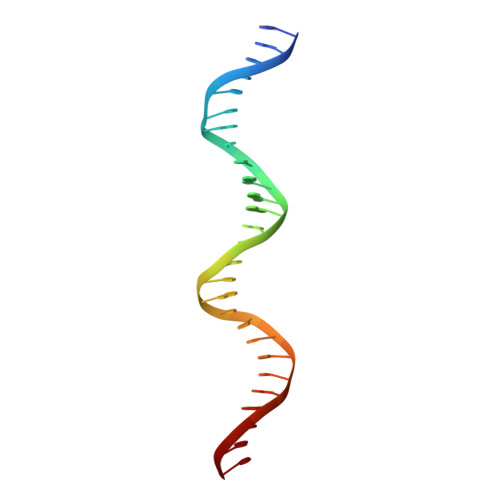
Structural studies of Ets-1/Pax5 complex formation on DNA.
Garvie, C.W., Hagman, J., Wolberger, C.(2001) Mol Cell 8: 1267-1276
- PubMed: 11779502
- DOI: https://doi.org/10.1016/s1097-2765(01)00410-5
- Primary Citation of Related Structures:
1K78, 1K79, 1K7A - PubMed Abstract:
Pax5 regulates the B cell-specific expression of the mb-1 gene together with members of the Ets family of transcriptional activators. The Ets proteins on their own bind poorly to the Pax5/Ets binding site, but can be recruited to the site by cooperative interactions with Pax5. The structure of the ETS domain of Ets-1 and the paired domain of Pax5 bound to DNA reveals the molecular details of the selective recruitment of different Ets proteins by Pax5. Comparison with structures of Ets-1 alone bound to both high- and low-affinity DNA sites reveals that Pax5 alters the Ets-1 contacts with DNA. The ability of one protein to alter the DNA sequence-specific contacts of another provides a general mechanism for combinatorial regulation of transcription.
- Department of Biophysics and Biophysical Chemistry, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore, MD 21205, USA.
Organizational Affiliation: