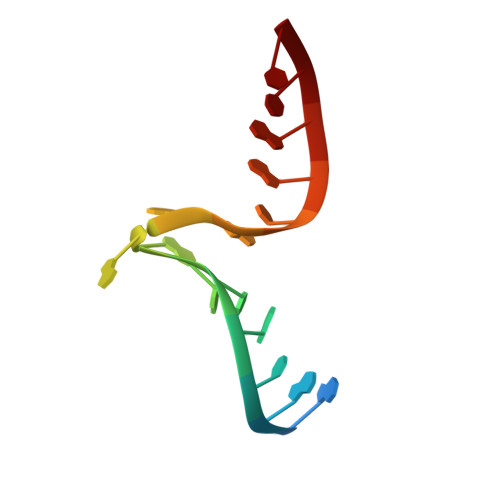Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Chao, J.A., Williamson, J.R.(2004) Structure 12: 1165-1176
- PubMed: 15242593
- DOI: https://doi.org/10.1016/j.str.2004.04.023
- Primary Citation of Related Structures:
1T0K - PubMed Abstract:
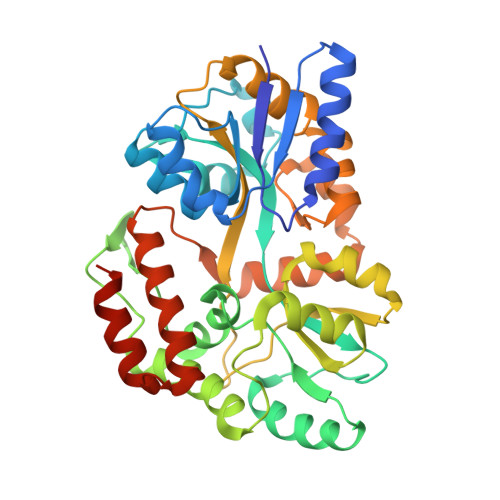
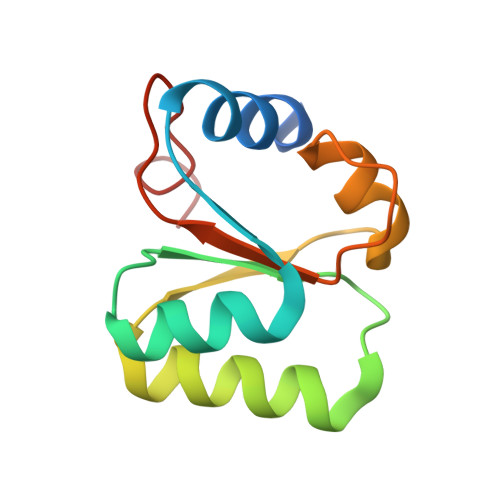
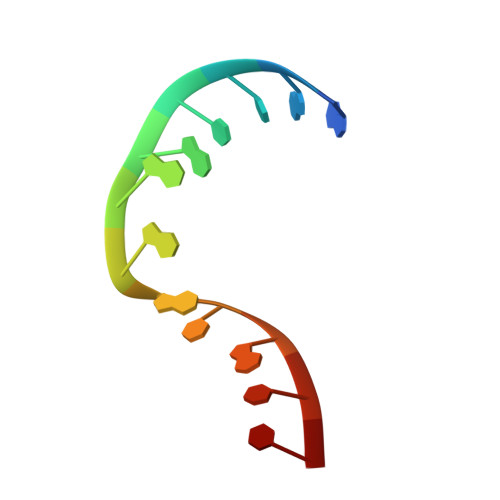
L30e, a Saccharomyces cervisiae ribosomal protein, regulates its own expression by binding to a purine-rich asymmetric internal loop located in both its pre-mRNA and mature mRNA. A crystal structure of an MBP-L30e fusion protein in complex with an RNA containing the pre-mRNA regulatory site was solved at 3.24 A. Interestingly, the structure of the RNA differed from that observed in a previously determined NMR structure of the complex. Analysis of the NMR data led to the identification of a single imino proton resonance in the internal loop that had been incorrectly assigned and was principally responsible for the erroneous RNA structure. A structure refinement was performed using both the X-ray diffraction data and the NMR-derived distance and angle restraints. The joint NMR and X-ray refinement resulted in improved stereochemistry and lower crystallographic R factors. The RNA internal loop of the MBP-L30e-mRNA complex adopts the canonical K-turn fold.
- Department of Molecular Biology, The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: