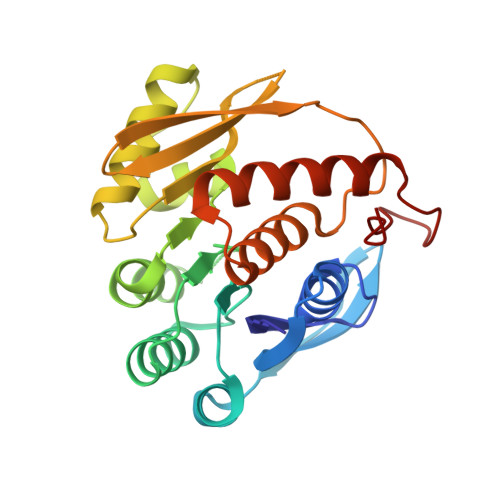
Crystal structure of 4-amino-5-hydroxymethyl-2-methyl pyrimidine phosphate kinase (HMPP kinase) from Thermus thermophilus HB8.
Kambaru, A., Jos, S., Lobov, I., Bagautdinov, B., Padavattan, S.(2025) Eur Biophys J 54: 247-256
- PubMed: 40483615
- DOI: https://doi.org/10.1007/s00249-025-01760-0
- Primary Citation of Related Structures:
1UB0 - PubMed Abstract:
The thiamine (vitamin B1) biosynthesis pathway is essential for most prokaryotes and some eukaryotes, including yeasts and plants. The 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (HMPP kinase), encoded by the thiD gene, catalyzes two phosphorylation reactions involving intermediates in this pathway, ultimately producing thiamine pyrophosphate, the active form of thiamine. Here, we present the crystal structure of HMPP kinase from Thermus thermophilus HB8 (TtHMPPK), resolved at a resolution of 2.05 Å. The asymmetric unit of the TtHMPPK structure includes one protomer, though it functions as a homodimer in its active form, like the HMPP kinase from Salmonella typhimurium. The TtHMPPK protomer is an α/β protein featuring nine β-sheets flanked by eight structurally conserved α-helices, which are characteristic of the ribokinase family. The structure reveals a Rossmann β-α-β motif, commonly found in nucleotide-binding proteins. Structural analysis of TtHMPPK, compared to the Salmonella typhimurium HMPP kinase, indicates that Ala16, Thr40, Gln42, Ala78, and Val105 are active site residues involved in catalysis. The structural studies suggest that TtHMPPK belongs to the ribokinase superfamily and exhibits structural similarities with an enzyme containing a Rossmann-like structural motif (RLM). This Rossmann fold enables HMPP kinase to catalyze the phosphorylation of HMPP, a critical step in thiamine production.
- Department of Biophysics, National Institute of Mental Health and Neurosciences, Bangalore, 560029, India.
Organizational Affiliation: