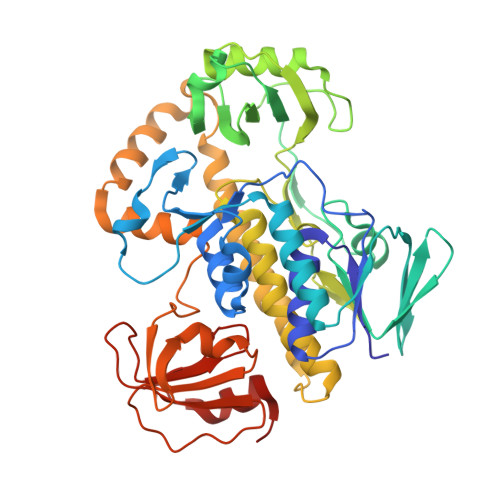
Structural Insights into a Flavin-Dependent [4 + 2] Cyclase that Catalyzes trans-Decalin Formation in Pyrroindomycin Biosynthesis.
Zheng, Q., Gong, Y., Guo, Y., Zhao, Z., Wu, Z., Zhou, Z., Chen, D., Pan, L., Liu, W.(2018) Cell Chem Biol 25: 718-727.e3
- PubMed: 29657086
- DOI: https://doi.org/10.1016/j.chembiol.2018.03.007
- Primary Citation of Related Structures:
5XGV - PubMed Abstract:
Here, we provide structural insights into PyrE3, a flavin-dependent [4 + 2] cyclase that catalyzes trans-decalin formation in the biosynthesis of pyrroindomycins. PyrE3 shares an architecture/domain organization head-to-tail similarity with the members of the family of para-hydroxybenzoate hydroxylase (pHBH)-fold monooxygenases, and possesses a flavin adenine dinucleotide (FAD)-binding domain, a middle domain, and a C-terminal thioredoxin-like domain. The FAD-binding domain forms a central hub of the protein structure, and binds with FAD in a "closed" conformation of pHBH-fold family monooxygenases known for their highly dynamic catalytic processes. FAD plays an essential structural role in PyrE3, where it is amenable to redox change; however, redox change has little effect on [4 + 2] cyclization activity. PyrE3 appears to selectively accommodate a tetramate-containing, linear polyene intermediate in a highly positively charged pocket, which is located at the interface between the FAD-binding domain and the middle domain, and can accelerate trans-decalin formation likely through an endo-selective [4 + 2] transition state.
- State Key Laboratory of Bioorganic and Natural Products Chemistry, Center for Excellence in Molecular Sciences, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, 345 Lingling Road, Shanghai 200032, China.
Organizational Affiliation: