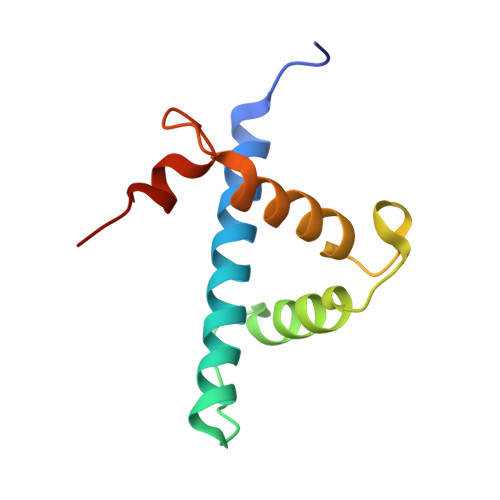
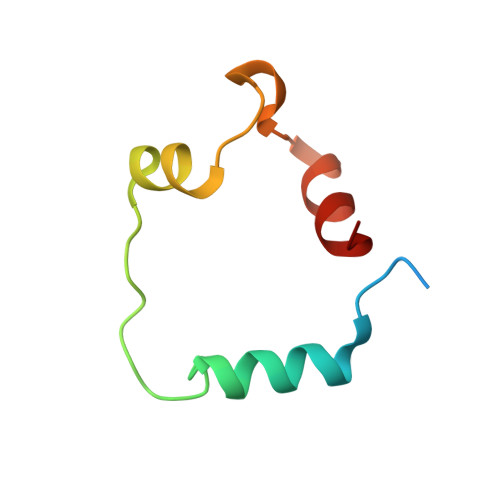
The molecular basis of allostery in a facilitated dissociation process.
Appling, F.D., Berlow, R.B., Stanfield, R.L., Dyson, H.J., Wright, P.E.(2021) Structure 29: 1327-1338.e5
- PubMed: 34520739
- DOI: https://doi.org/10.1016/j.str.2021.07.011
- Primary Citation of Related Structures:
7LVS - PubMed Abstract:
Facilitated dissociation provides a mechanism by which high-affinity complexes can be rapidly disassembled. The negative feedback regulator CITED2 efficiently downregulates the hypoxic response by displacing the hypoxia-inducible transcription factor HIF-1α from the TAZ1 domain of the transcriptional coactivators CREB-binding protein (CBP) and p300. Displacement occurs by a facilitated dissociation mechanism involving a transient ternary intermediate formed by binding of the intrinsically disordered CITED2 activation domain to the TAZ1:HIF-1α complex. The short lifetime of the intermediate precludes straightforward structural investigations. To obtain insights into the molecular determinants of facilitated dissociation, we model the ternary intermediate by generating a fusion peptide composed of the primary CITED2 and HIF-1α binding motifs. X-ray crystallographic and NMR studies of the fusion peptide complex reveal TAZ1-mediated negative cooperativity that results in nearly mutually exclusive binding of specific CITED2 and HIF-1α interaction motifs, providing molecular-level insights into the allosteric switch that terminates the hypoxic response.
- Department of Integrative Structural and Computational Biology and Skaggs Institute of Chemical Biology, Scripps Research, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: