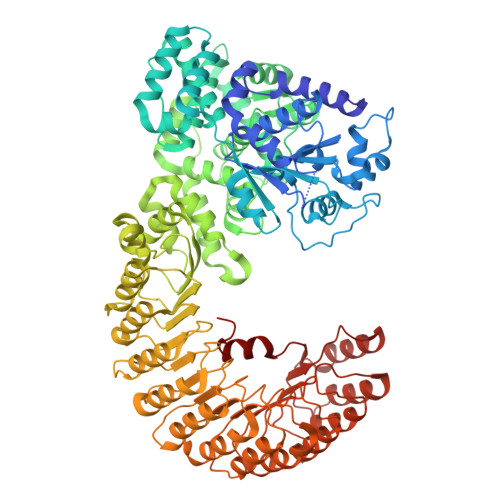
The structure of NLRP9 reveals a unique C-terminal region with putative regulatory function.
Kamitsukasa, Y., Nakano, K., Murakami, K., Hirata, K., Yamamoto, M., Shimizu, T., Ohto, U.(2022) FEBS Lett 596: 876-885
- PubMed: 35090055
- DOI: https://doi.org/10.1002/1873-3468.14302
- Primary Citation of Related Structures:
7WBT, 7WBU - PubMed Abstract:
Nucleotide-binding and oligomerisation domain-like receptors (NLRs) can form inflammasomes that activate caspase-1 and pro-interleukin-1β and induce pyroptosis. NLR family pyrin domain-containing 9 (NLRP9) forms an inflammasome and activates innate immune responses during virus infection, but little is known about this process. Here, we report the crystal and cryo-electron microscopy structures of NLRP9 in an ADP-bound state, revealing inactive and closed conformations of NLRP9 and its similarities to other structurally characterised NLRs. Moreover, we found a C-terminal region interacting with the concave surface of the leucine-rich repeat domain of NLRP9. This region is unique among NLRs and might be involved in the specific function of NLRP9. These data provide the structural basis for understanding the mechanism of NLRP9 regulation and activation.
- Graduate School of Pharmaceutical Sciences, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: