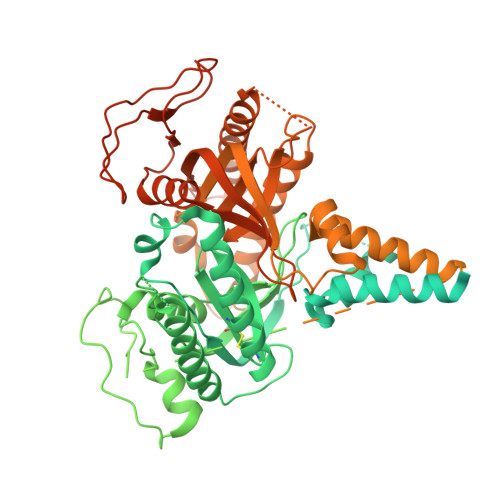
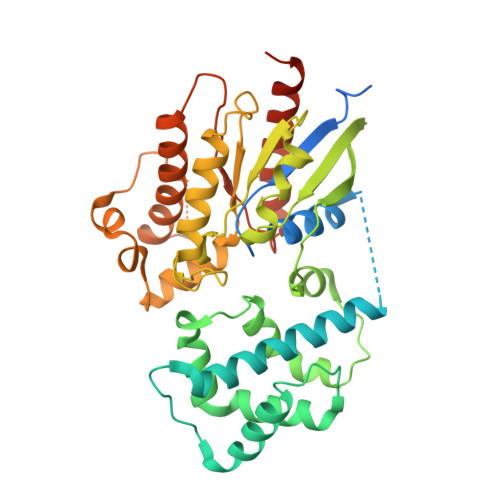
Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.
Khanppnavar, B., Schuster, D., Lavriha, P., Uliana, F., Ozel, M., Mehta, V., Leitner, A., Picotti, P., Korkhov, V.M.(2024) EMBO Rep 25: 1513-1540
- PubMed: 38351373
- DOI: https://doi.org/10.1038/s44319-024-00076-y
- Primary Citation of Related Structures:
8BUZ, 8BV5, 9A3T - PubMed Abstract:
Membrane adenylyl cyclase AC8 is regulated by G proteins and calmodulin (CaM), mediating the crosstalk between the cAMP pathway and Ca 2+ signalling. Despite the importance of AC8 in physiology, the structural basis of its regulation by G proteins and CaM is not well defined. Here, we report the 3.5 Å resolution cryo-EM structure of the bovine AC8 bound to the stimulatory Gαs protein in the presence of Ca 2+ /CaM. The structure reveals the architecture of the ordered AC8 domains bound to Gαs and the small molecule activator forskolin. The extracellular surface of AC8 features a negatively charged pocket, a potential site for unknown interactors. Despite the well-resolved forskolin density, the captured state of AC8 does not favour tight nucleotide binding. The structural proteomics approaches, limited proteolysis and crosslinking mass spectrometry (LiP-MS and XL-MS), allowed us to identify the contact sites between AC8 and its regulators, CaM, Gαs, and Gβγ, as well as to infer the conformational changes induced by these interactions. Our results provide a framework for understanding the role of flexible regions in the mechanism of AC regulation.
Organizational Affiliation:
Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institute, Villigen, Switzerland.