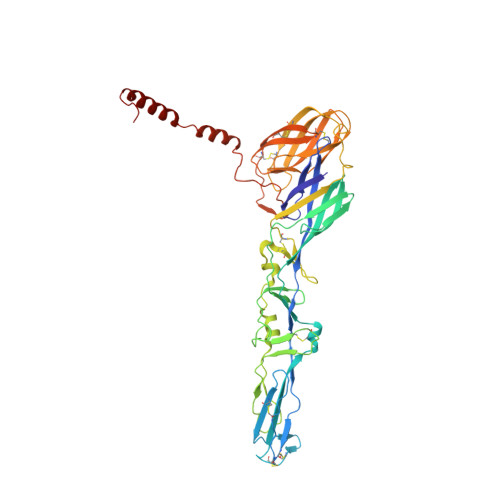
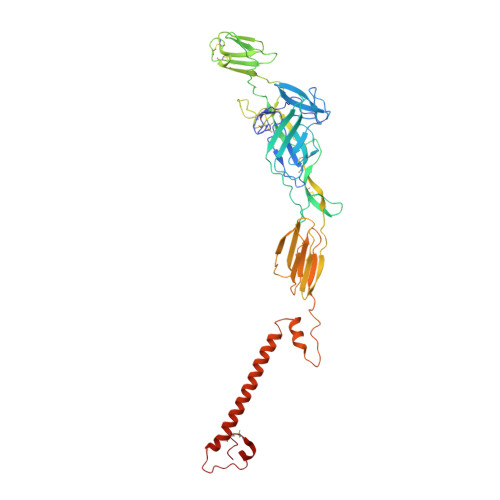
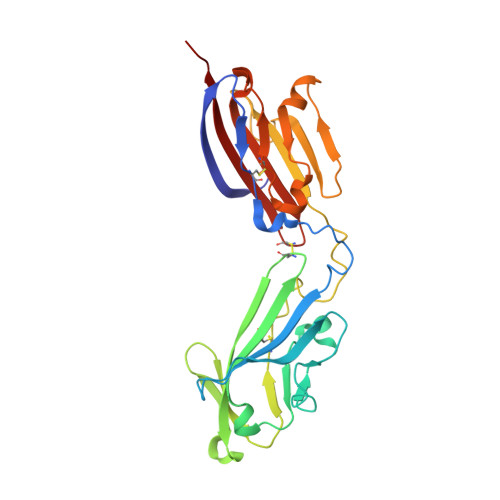
Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Zimmerman, O., Zimmerman, M.I., Raju, S., Nelson, C.A., Errico, J.M., Madden, E.A., Holmes, A.C., Hassan, A.O., VanBlargan, L.A., Kim, A.S., Adams, L.J., Basore, K., Whitener, B.M., Palakurty, S., Davis-Adams, H.G., Sun, C., Gilliland Jr., T., Earnest, J.T., Ma, H., Ebel, G.D., Zmasek, C., Scheuermann, R.H., Klimstra, W.B., Fremont, D.H., Diamond, M.S.(2023) Cell 186: 4818-4833.e25
- PubMed: 37804831
- DOI: https://doi.org/10.1016/j.cell.2023.09.007
- Primary Citation of Related Structures:
8DAN, 8DAQ, 8EWF, 8SQN - PubMed Abstract:
MXRA8 is a receptor for chikungunya (CHIKV) and other arthritogenic alphaviruses with mammalian hosts. However, mammalian MXRA8 does not bind to alphaviruses that infect humans and have avian reservoirs. Here, we show that avian, but not mammalian, MXRA8 can act as a receptor for Sindbis, western equine encephalitis (WEEV), and related alphaviruses with avian reservoirs. Structural analysis of duck MXRA8 complexed with WEEV reveals an inverted binding mode compared with mammalian MXRA8 bound to CHIKV. Whereas both domains of mammalian MXRA8 bind CHIKV E1 and E2, only domain 1 of avian MXRA8 engages WEEV E1, and no appreciable contacts are made with WEEV E2. Using these results, we generated a chimeric avian-mammalian MXRA8 decoy-receptor that neutralizes infection of multiple alphaviruses from distinct antigenic groups in vitro and in vivo. Thus, different alphaviruses can bind MXRA8 encoded by different vertebrate classes with distinct engagement modes, which enables development of broad-spectrum inhibitors.
- Department of Medicine, Washington University School of Medicine, Saint Louis, MO 63110, USA.
Organizational Affiliation: