Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Lu, D., Chen, Y., Jiang, M., Tan, S.G., Chai, Y., Gao, G.F.(2023) Nat Commun
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MHC class I antigen (Fragment) | 274 | Homo sapiens | Mutation(s): 2 Gene Names: HLA-A | 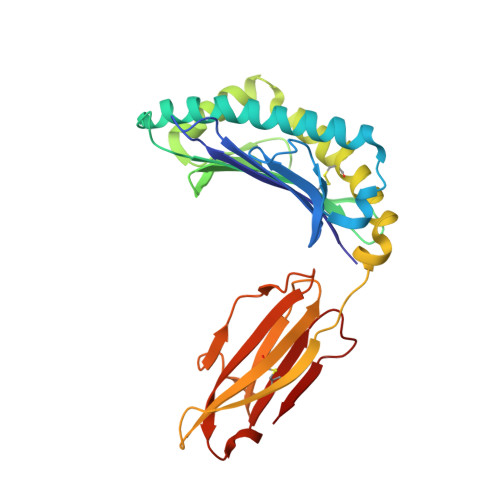 | |
UniProt | |||||
Find proteins for U5YJJ6 (Homo sapiens) Explore U5YJJ6 Go to UniProtKB: U5YJJ6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | U5YJJ6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 99 | Homo sapiens | Mutation(s): 0 Gene Names: B2M | 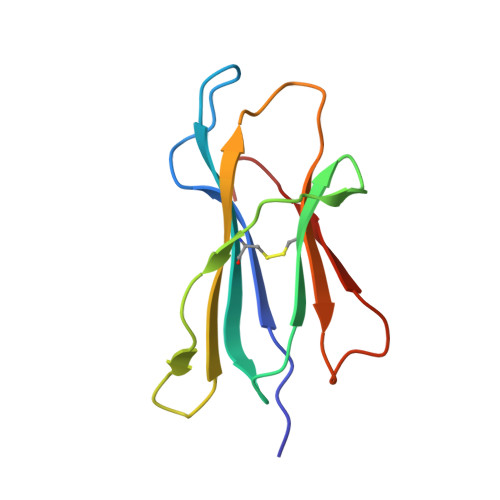 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| peptide KRAS-G12V-9 | 9 | Homo sapiens | Mutation(s): 0 EC: 3.6.5.2 | 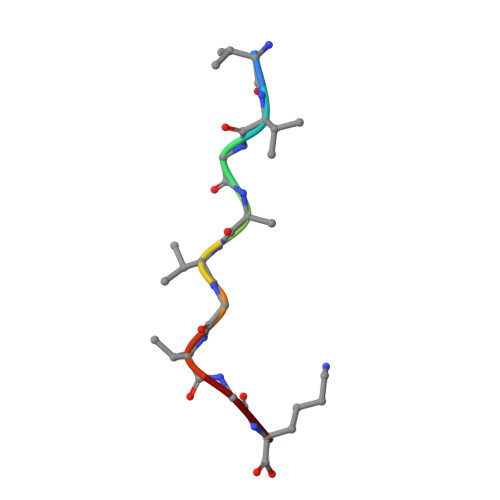 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01116 (Homo sapiens) Explore P01116 Go to UniProtKB: P01116 | |||||
PHAROS: P01116 GTEx: ENSG00000133703 | |||||
Entity Groups | |||||
| UniProt Group | P01116 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TCR alpha chain | 199 | Mus musculus | Mutation(s): 0 | 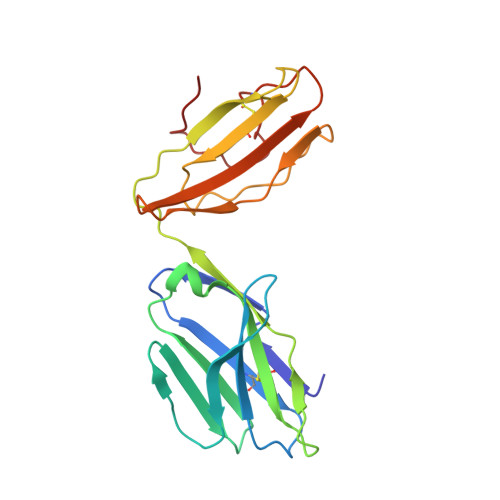 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TCR beta chain | 242 | Mus musculus | Mutation(s): 0 | 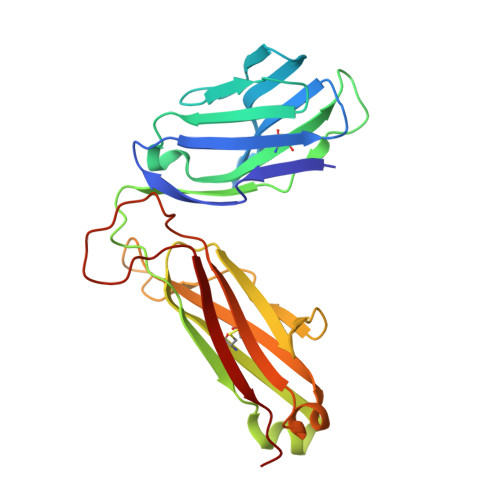 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 156.864 | α = 77.37 |
| b = 156.889 | β = 78.13 |
| c = 191.966 | γ = 82.27 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data reduction |
| PHENIX | refinement |
| Coot | model building |
| HKL-2000 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 2022YFC2302900 |
| National Natural Science Foundation of China (NSFC) | China | 2021YFC2301400 |
| National Natural Science Foundation of China (NSFC) | China | 92169208 |
| National Natural Science Foundation of China (NSFC) | China | 32222031 |