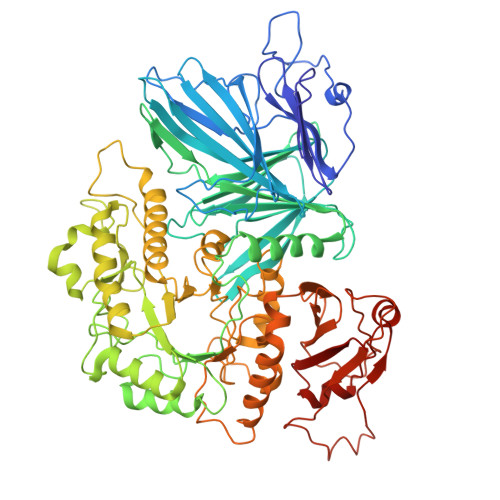
Activity-based metaproteomics driven discovery and enzymological characterization of potential alpha-galactosidases in the mouse gut microbiome.
Jiang, J., Czuchry, D., Ru, Y., Peng, H., Shen, J., Wang, T., Zhao, W., Chen, W., Sui, S.F., Li, Y., Li, N.(2024) Commun Chem 7: 184-184
- PubMed: 39152233
- DOI: https://doi.org/10.1038/s42004-024-01273-5
- Primary Citation of Related Structures:
8K1A, 8K7U, 8K7V - PubMed Abstract:
The gut microbiota offers an extensive resource of enzymes, but many remain uncharacterized. To distinguish the activities of similar annotated proteins and mine the potentially applicable ones in the microbiome, we applied an effective Activity-Based Metaproteomics (ABMP) strategy using a specific activity-based probe (ABP) to screen the entire gut microbiome for directly discovering active enzymes and their potential applications, not for exploring host-microbiome interactions. By using an activity-based cyclophellitol aziridine probe specific to α-galactosidases (AGAL), we successfully identified and characterized several gut microbiota enzymes possessing AGAL activities. Cryo-electron microscopy analysis of a newly characterized enzyme (AGLA5) revealed the covalent binding conformations between the AGAL5 active site and the cyclophellitol aziridine ABP, which could provide insights into the enzyme's catalytic mechanism. The four newly characterized AGALs have diverse potential activities, including raffinose family oligosaccharides (RFOs) hydrolysis and enzymatic blood group transformation. Collectively, we present a ABMP platform that facilitates gut microbiota AGALs discovery, biochemical activity annotations and potential industrial or biopharmaceutical applications.
- Institute for Inheritance-Based Innovation of Chinese Medicine, School of Pharmacy, Shenzhen University Medical School, Shenzhen University, Shenzhen, 518055, China.
Organizational Affiliation: