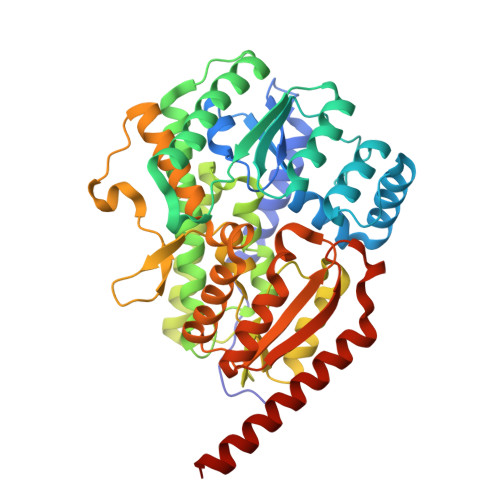
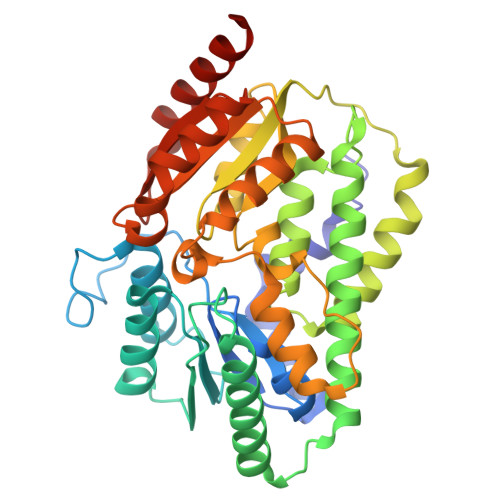
Enzymatic Birch reduction via hydrogen atom transfer at [4Fe-4S]-OH 2 and [8Fe-9S] clusters.
Fuchs, J., Fernandez-Arevalo, U., Demmer, U., Diaz, E., Ullmann, G.M., Pierik, A.J., Ermler, U., Boll, M.(2025) Nat Commun 16: 3236-3236
- PubMed: 40185728
- DOI: https://doi.org/10.1038/s41467-025-58418-w
- Primary Citation of Related Structures:
8S02, 8S1T, 8S2R - PubMed Abstract:
The alkali metal- and ammonia-dependent Birch reduction is the classical synthetic method for achieving dihydro additions to arenes, typically yielding 1,4-cyclodienes. A mild biological alternative to this process are 1,5-dienoyl-coenzyme A (CoA)-forming class I and II benzoyl-CoA reductases (BCRs), widely abundant key enzymes in the biodegradation of aromatic compounds at anoxic environments. To obtain a comprehensive mechanistic understanding of class I BCR catalysis, we produced the active site subunits from a denitrifying bacterium and determined the X-ray structure of its substrate and product complexes at 1.4 Å revealing non-canonical double-cubane [8Fe-9S] and active site aqua-[4Fe-4S] clusters. Together with kinetic, spectroscopic and QM/MM studies, we provide evidence for a radical mechanism with a [4Fe-4S] cluster-bound water molecule acting as hydrogen atom and electron donor at potentials beyond the biological redox window. An analogous Birch-like radical mechanism is applied by class II BCRs with the catalytic water bound to a tungsten-bis-metallopterin cofactor. The use of activated, metal-bound water ligands as hydrogen atom donor serves as a basic blueprint for future enzymatic or biomimetic Birch reduction processes.
- Faculty of Biology - Microbiology, University of Freiburg, 79104, Freiburg, Germany.
Organizational Affiliation: