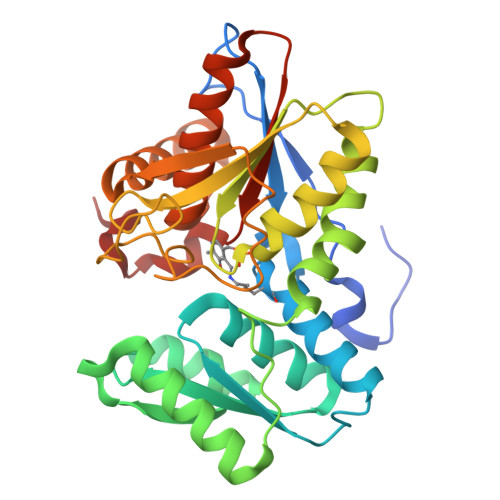
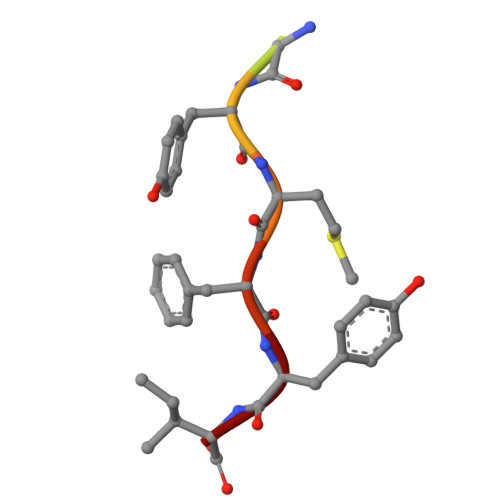
Identification of Cysteine Metabolism Regulator (CymR)-Derived Pentapeptides as Nanomolar Inhibitors of Staphylococcus aureus O -Acetyl-l-serine Sulfhydrylase (CysK).
Pederick, J.L., Vandborg, B.C., George, A., Bovermann, H., Boyd, J.M., Freundlich, J.S., Bruning, J.B.(2025) ACS Infect Dis 11: 238-248
- PubMed: 39705018
- DOI: https://doi.org/10.1021/acsinfecdis.4c00832
- Primary Citation of Related Structures:
8SRT, 8SRU, 8SRV, 8SRW, 8T2C - PubMed Abstract:
The pathway of bacterial cysteine biosynthesis is gaining traction for the development of antibiotic adjuvants. Bacterial cysteine biosynthesis is generally facilitated by two enzymes possessing O -acetyl-l-serine sulfhydrylases (OASS), CysK and CysM. In Staphylococcus aureus , there exists a single OASS homologue, Sa CysK. Knockout of Sa CysK was found to increase sensitivity to oxidative stress, making it a relevant target for inhibitor development. Sa CysK also forms two functional complexes via interaction with the preceding enzyme in the pathway serine acetyltransferase (CysE) or the transcriptional regulator of cysteine metabolism (CymR). These interactions occur through insertion of a C-terminal peptide of CysE or CymR into the active site of Sa CysK, inhibiting OASS activity, and therefore represent an excellent starting point for developing Sa CysK inhibitors. Here, we detail the characterization of CysE and CymR-derived C-terminal peptides as inhibitors of Sa CysK. Using a combination of X-ray crystallography, surface plasmon resonance, and enzyme inhibition assays, it was determined that the CymR-derived decapeptide forms extensive interactions with Sa CysK and acts as a potent inhibitor ( K D = 25 nM; IC 50 = 180 nM), making it a promising lead for the development of Sa CysK inhibitors. To understand the determinants of this high-affinity interaction, the structure-activity relationships of 16 rationally designed peptides were also investigated. This identified that the C-terminal pentapeptide of CymR facilitates the high-affinity interaction with Sa CysK and that subtle structural modification of the pentapeptide is possible without impacting potency. Ultimately, this work identified CymR pentapeptides as a promising scaffold for the development of antibiotic adjuvants targeting Sa CysK.
- Institute for Photonics and Advanced Sensing (IPAS), School of Biological Sciences, The University of Adelaide, Adelaide, South Australia 5005, Australia.
Organizational Affiliation: