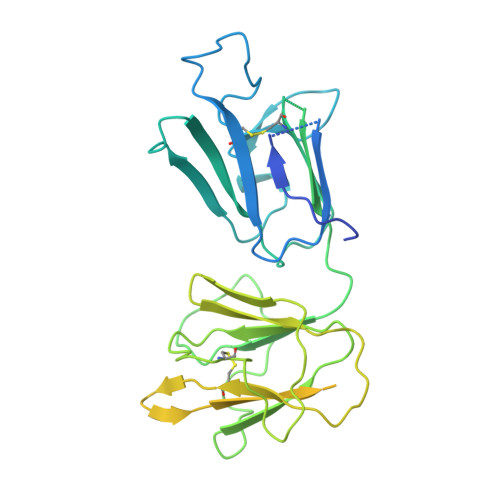
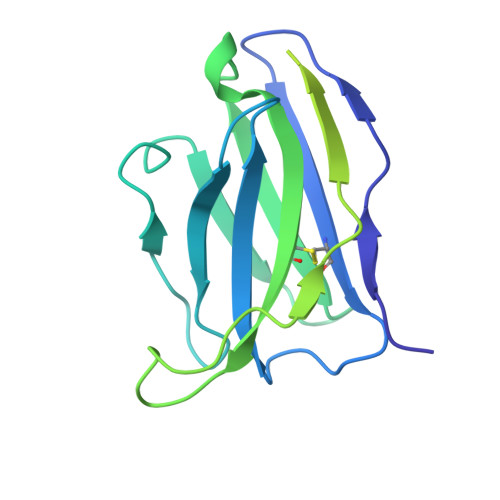
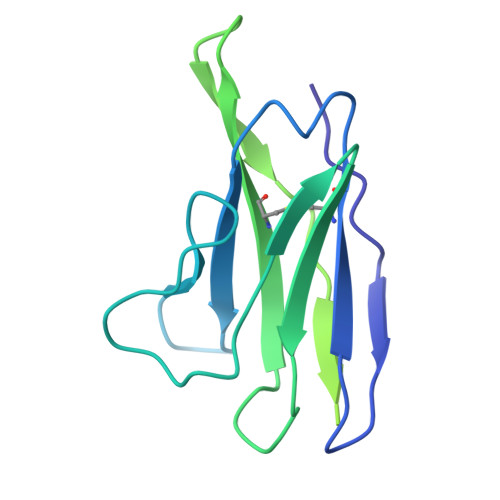
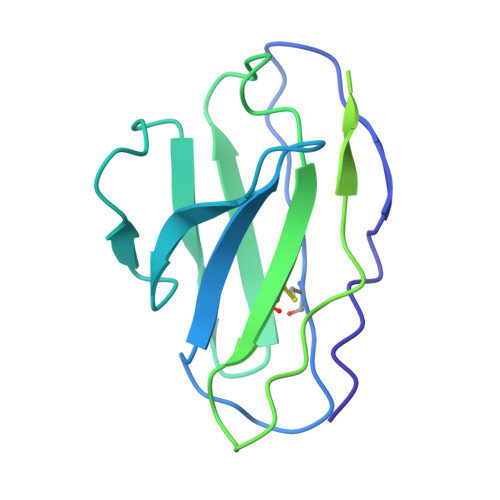
Structural elucidation of full-length Pfs48/45 in complex with potent monoclonal antibodies isolated from a naturally exposed individual.
Kucharska, I., Ivanochko, D., Hailemariam, S., Inklaar, M.R., Kim, H.R., Teelen, K., Stoter, R., van de Vegte-Bolmer, M., van Gemert, G.J., Semesi, A., McLeod, B., Ki, A., Lee, W.K., Rubinstein, J.L., Jore, M.M., Julien, J.P.(2025) Nat Struct Mol Biol 32: 1396-1407
- PubMed: 40404982
- DOI: https://doi.org/10.1038/s41594-025-01532-6
- Primary Citation of Related Structures:
8U1P, 8U70 - PubMed Abstract:
Biomedical interventions that block the transmission of Plasmodium falciparum (Pf) from humans to mosquitoes may be critical for malaria elimination. Pfs48/45, a gamete-surface protein essential for Pf development in the mosquito midgut, is a target of clinical-stage transmission-blocking vaccines and monoclonal antibodies (mAbs) that disrupt Pf transmission to mosquitoes. Antibodies directed to domain 3 of Pfs48/45 have been structurally and functionally described; however, in-depth information about other inhibitory epitopes on Pfs48/45 is currently limited. Here, we present a cryo-electron microscopy structure of full-length Pfs48/45 in complex with potent human mAbs targeting all three domains. Our data indicate that although Pfs48/45 domains 1 and 2 are rigidly coupled, there is substantial conformational flexibility between domains 2 and 3. Characterization of mAbs against domain 1 revealed the presence of a conformational epitope class that is largely conserved across Pf field isolates and is associated with recognition by potent antibodies. Our study provides insights into epitopes across full-length Pfs48/45 and has implications for the design of next-generation malaria interventions.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Ontario, Canada.
Organizational Affiliation: