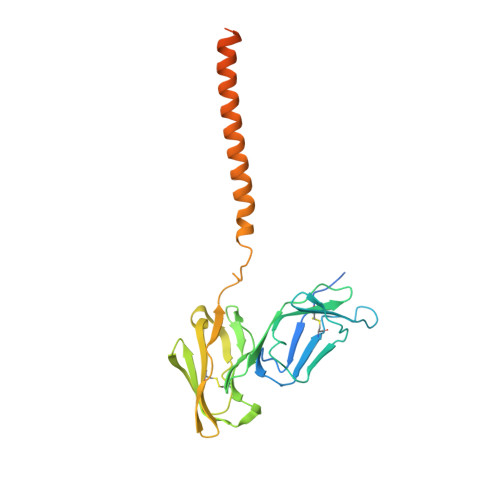
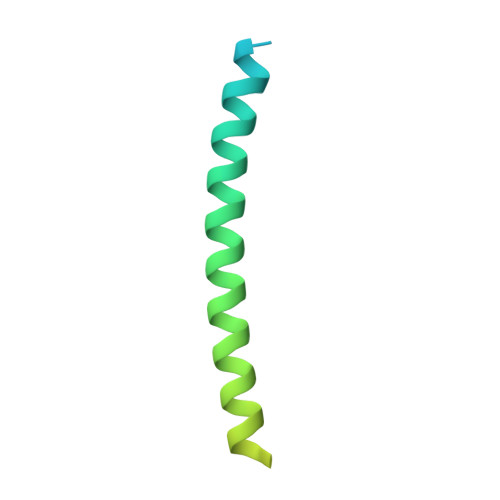
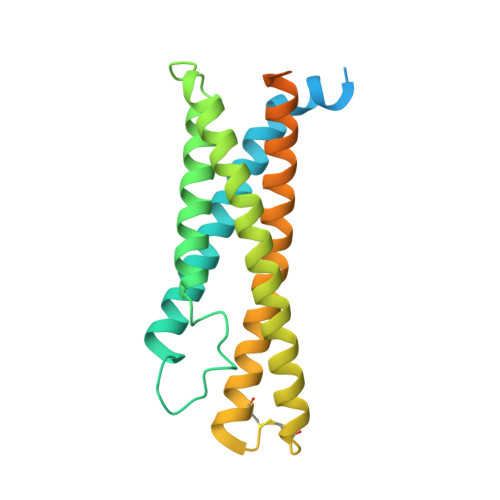
Architecture of the high-affinity immunoglobulin E receptor.
Zhang, Z., Yui, M., Ohto, U., Shimizu, T.(2024) Sci Signal 17: eadn1303-eadn1303
- PubMed: 39656861
- DOI: https://doi.org/10.1126/scisignal.adn1303
- Primary Citation of Related Structures:
8K7R, 8K7S, 8K7T, 8YRJ - PubMed Abstract:
The high-affinity immunoglobulin E (IgE) receptor (FcεRI) drives type I hypersensitivity in response to allergen-specific IgE. FcεRI is a multimeric complex typically composed of one α, one β, and two disulfide-linked γ subunits. The α subunit binds to the fragment crystallizable (Fc) region of IgE (Fcε), whereas the β and γ subunits mediate signaling through their intracellular immunoreceptor tyrosine-based activation motifs (ITAMs). Here, we report cryo-electron microscopy (cryo-EM) structures of the apo state of FcεRI and of FcεRI bound to Fcε. At the transmembrane domain (TMD), the α and γ subunits associate to form a tightly packed, three-helix bundle (αγ 2 bundle) with pseudo-threefold symmetry through extensive hydrophobic and polar interactions. The αγ 2 bundle further assembles with the β subunit to complete the TMD, from which multiple ITAMs might extend into the cytoplasm for downstream signaling. The apo mouse FcεRI essentially forms an identical structure to that of the Fcε-bound sensitized form, suggesting that the binding of Fcε to FcεRI does not alter the overall conformation of the receptor. Furthermore, the juxtamembrane interaction between the extracellular domains (ECDs) of mouse FcεRIα and FcεRIβ is not observed between their human counterparts, which implies potential species-specific differences in receptor stability and activation. Our findings provide a framework for understanding the general structural principles underlying Fc receptor assembly, the signaling mechanism underlying type I hypersensitivity, and the design of efficient antiallergic therapeutics.
- Graduate School of Pharmaceutical Sciences, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan.
Organizational Affiliation: