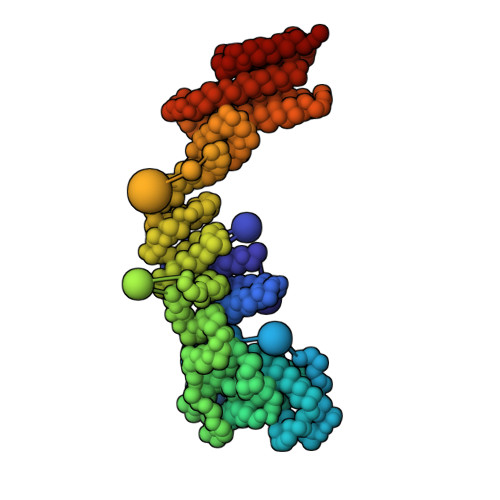
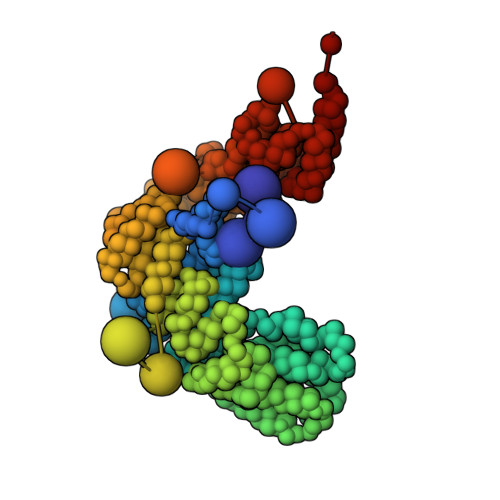
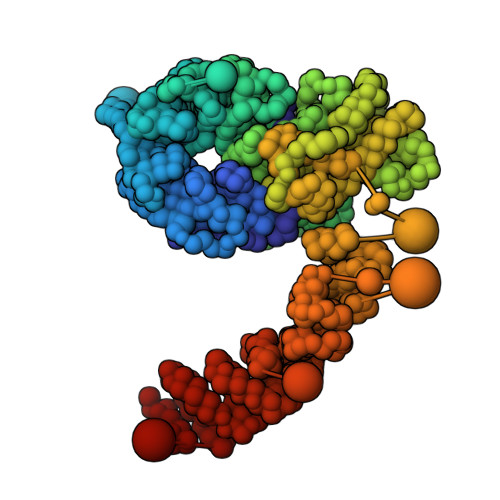
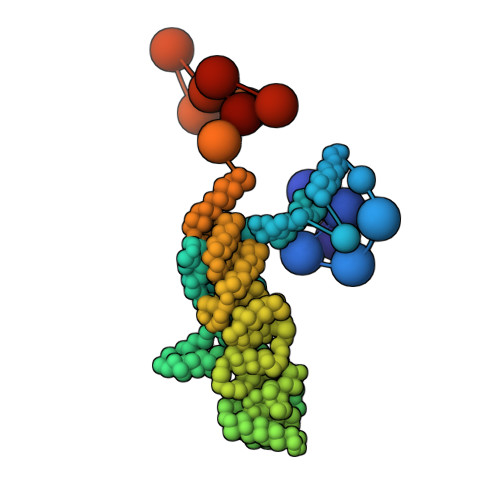
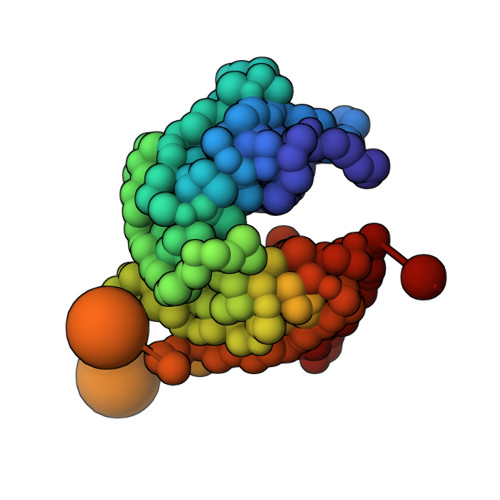
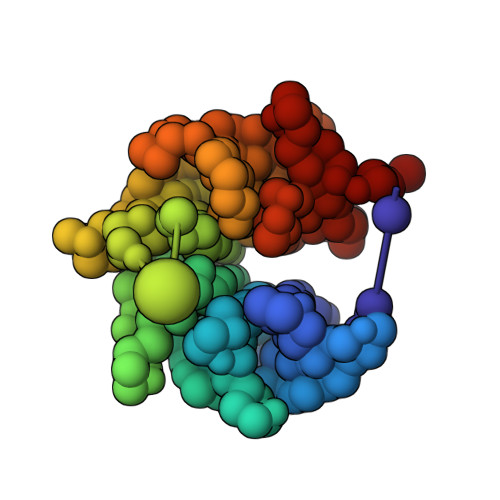
Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex.
Shi, Y., Fernandez-Martinez, J., Tjioe, E., Pellarin, R., Kim, S.J., Williams, R., Schneidman-Duhovny, D., Sali, A., Rout, M.P., Chait, B.T.(2014) Mol Cell Proteomics 13: 2927-2943
- PubMed: 25161197
- DOI: https://doi.org/10.1074/mcp.M114.041673
- Primary Citation of Related Structures:
8ZZ1 - PubMed Abstract:
Most cellular processes are orchestrated by macromolecular complexes. However, structural elucidation of these endogenous complexes can be challenging because they frequently contain large numbers of proteins, are compositionally and morphologically heterogeneous, can be dynamic, and are often of low abundance in the cell. Here, we present a strategy for the structural characterization of such complexes that has at its center chemical cross-linking with mass spectrometric readout. In this strategy, we isolate the endogenous complexes using a highly optimized sample preparation protocol and generate a comprehensive, high-quality cross-linking dataset using two complementary cross-linking reagents. We then determine the structure of the complex using a refined integrative method that combines the cross-linking data with information generated from other sources, including electron microscopy, X-ray crystallography, and comparative protein structure modeling. We applied this integrative strategy to determine the structure of the native Nup84 complex, a stable hetero-heptameric assembly (∼ 600 kDa), 16 copies of which form the outer rings of the 50-MDa nuclear pore complex (NPC) in budding yeast. The unprecedented detail of the Nup84 complex structure reveals previously unseen features in its pentameric structural hub and provides information on the conformational flexibility of the assembly. These additional details further support and augment the protocoatomer hypothesis, which proposes an evolutionary relationship between vesicle coating complexes and the NPC, and indicates a conserved mechanism by which the NPC is anchored in the nuclear envelope.
- From the ‡Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, The Rockefeller University, New York, New York 10065;
Organizational Affiliation: