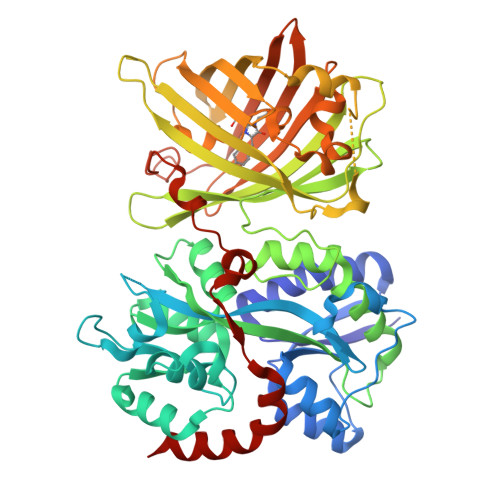
iGABASnFR2: Improved genetically encoded protein sensors of GABA
Kolb, I., Hasseman, J.P., Matsumoto, A., Jensen, T.P., Kopach, O., Arthur, B.J., Zhang, Y., Tsang, A., Reep, D., Tsegaye, G., Zheng, J., Patel, R.H., Looger, L.L., Marvin, J.S., Korff, W.L., Rusakov, D.A., Yonehara, K., Turner, G.C.(2025) Elife