Ni,Fe-CODH : Ti(III)-reduced pH 8.0
Basak, Y., Jeoung, J.-H., Dobbek, H.(2025) Nature Catalysis
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Carbon monoxide dehydrogenase 2 | A [auth X] | 636 | Carboxydothermus hydrogenoformans Z-2901 | Mutation(s): 0 Gene Names: cooS2, cooSII, CHY_0085 EC: 1.2.7.4 | 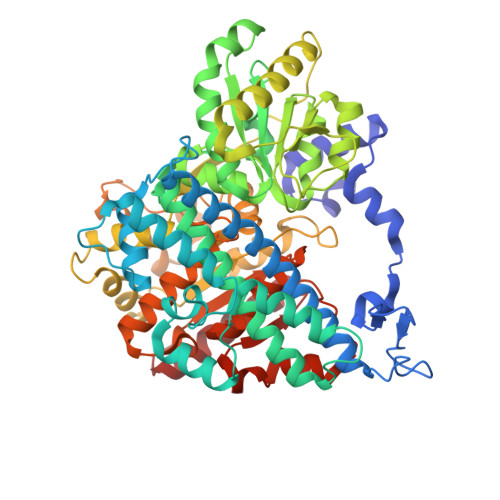 |
UniProt | |||||
Find proteins for Q9F8A8 (Carboxydothermus hydrogenoformans (strain ATCC BAA-161 / DSM 6008 / Z-2901)) Explore Q9F8A8 Go to UniProtKB: Q9F8A8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9F8A8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 9 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SF4 Query on SF4 | B [auth X] | IRON/SULFUR CLUSTER Fe4 S4 LJBDFODJNLIPKO-UHFFFAOYSA-N |  | ||
| F3S (Subject of Investigation/LOI) Query on F3S | D [auth X] | FE3-S4 CLUSTER Fe3 S4 FCXHZBQOKRZXKS-UHFFFAOYSA-N |  | ||
| FES Query on FES | C [auth X] | FE2/S2 (INORGANIC) CLUSTER Fe2 S2 NIXDOXVAJZFRNF-UHFFFAOYSA-N |  | ||
| TRS Query on TRS | I [auth X] | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL C4 H12 N O3 LENZDBCJOHFCAS-UHFFFAOYSA-O |  | ||
| PEG Query on PEG | J [auth X], K [auth X] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| NI (Subject of Investigation/LOI) Query on NI | F [auth X] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| FE (Subject of Investigation/LOI) Query on FE | E [auth X] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CO2 (Subject of Investigation/LOI) Query on CO2 | H [auth X] | CARBON DIOXIDE C O2 CURLTUGMZLYLDI-UHFFFAOYSA-N |  | ||
| CMO (Subject of Investigation/LOI) Query on CMO | G [auth X] | CARBON MONOXIDE C O UGFAIRIUMAVXCW-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 112.25 | α = 90 |
| b = 74.97 | β = 111.4 |
| c = 71 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| MxCuBE | data collection |
| Coot | model building |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | DO 785/6-2 |
| Germanys Excellence Strategy | Germany | EXC 2008 - 390540038 - UniSysCat |